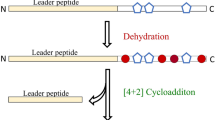
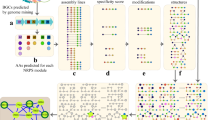
Abstract
Microbial natural products have played a key role in the development of clinical agents in nearly all therapeutic areas. Recent advances in genome sequencing have revealed that there is an incredible wealth of new polyketide and non-ribosomal peptide natural product diversity to be mined from genetic data. The diversity and complexity of polyketide and non-ribosomal peptide biosynthesis has required the development of unique bioinformatics tools to identify, annotate, and predict the structures of these natural products from their biosynthetic gene clusters. This review highlights and evaluates web-based bioinformatics tools currently available to the natural product community for genome mining to discover new polyketides and non-ribosomal peptides.


Similar content being viewed by others
References
Anand S, Prasad MVR, Yadav G et al (2010) SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res 38:W487–W496. doi:10.1093/nar/gkq340
Ansari MZ, Sharma J, Gokhale RS, Mohanty D (2008) In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites. BMC Bioinformatics 9:454. doi:10.1186/1471-2105-9-454
Beck BJ, Yoon YJ, Reynolds KA, Sherman DH (2002) The hidden steps of domain skipping: macrolactone ring size determination in the pikromycin modular polyketide synthase. Chem Biol 9:575–583
Blin K, Medema MH, Kazempour D et al (2013) antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41:W204–W212. doi:10.1093/nar/gkt449
Brikun IA, Reeves AR, Cernota WH et al (2004) The erythromycin biosynthetic gene cluster of Aeromicrobium erythreum. J Ind Microbiol Biotechnol 31:335–344. doi:10.1007/s10295-004-0154-5
Caboche S, Pupin M, Leclère V et al (2008) NORINE: a database of nonribosomal peptides. Nucleic Acids Res 36:D326–D331. doi:10.1093/nar/gkm792
Caffrey P (2003) Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. ChemBioChem 4:654–657. doi:10.1002/cbic.200300581
Callahan B, Thattai M, Shraiman BI (2009) Emergent gene order in a model of modular polyketide synthases. Proc Natl Acad Sci USA 106:19410–19415. doi:10.1073/pnas.0902364106
Challis GL, Ravel J, Townsend CA (2000) Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol 7:211–224
Chooi Y-H, Wang P, Fang J et al (2012) Discovery and characterization of a group of fungal polycyclic polyketide prenyltransferases. J Am Chem Soc 134:9428–9437. doi:10.1021/ja3028636
Clugston SL, Sieber SA, Marahiel MA, Walsh CT (2003) Chirality of peptide bond-forming condensation domains in nonribosomal peptide synthetases: the C5 domain of tyrocidine synthetase is a (D)C(L) catalyst. Biochemistry 42:12095–12104. doi:10.1021/bi035090+
Dereeper A, Guignon V, Blanc G et al (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi:10.1093/nar/gkn180
Donadio S, Monciardini P, Sosio M (2007) Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat Prod Rep 24:1073–1109. doi:10.1039/b514050c
Dunn BJ, Khosla C (2013) Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J R Soc Interface 10:20130297. doi:10.1098/rsif.2013.0297
Eddy SR (2004) What is a hidden Markov model? Nat Biotechnol 22:1315–1316. doi:10.1038/nbt1004-1315
Eustáquio AS, McGlinchey RP, Liu Y et al (2009) Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-l-methionine. Proc Natl Acad Sci USA 106:12295–12300. doi:10.1073/pnas.0901237106
Feng Z, Kallifidas D, Brady SF (2011) Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc Natl Acad Sci USA 108:12629–12634. doi:10.1073/pnas.1103921108
Finn RD, Mistry J, Tate J et al (2010) The Pfam protein families database. Nucleic Acids Res 38:D211–D222. doi:10.1093/nar/gkp985
Fritzsche K, Ishida K, Hertweck C (2008) Orchestration of discoid polyketide cyclization in the resistomycin pathway. J Am Chem Soc 130:8307–8316. doi:10.1021/ja800251m
Gaitatzis N, Silakowski B, Kunze B et al (2002) The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J Biol Chem 277:13082–13090. doi:10.1074/jbc.M111738200
Guo X, Liu T, Valenzano CR et al (2010) Mechanism and stereospecificity of a fully saturating polyketide synthase module: nanchangmycin synthase module 2 and its dehydratase domain. J Am Chem Soc 132:14694–14696. doi:10.1021/ja1073432
Hertweck C (2009) The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Eng 48:4688–4716. doi:10.1002/anie.200806121
Hur GH, Vickery CR, Burkart MD (2012) Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat Prod Rep 29:1074–1098. doi:10.1039/c2np20025b
Javidpour P, Das A, Khosla C, Tsai S-C (2011) Structural and biochemical studies of the hedamycin type II polyketide ketoreductase (HedKR): molecular basis of stereo- and regiospecificities. Biochemistry 50:7426–7439. doi:10.1021/bi2006866
Jenke-Kodama H, Sandmann A, Müller R, Dittmann E (2005) Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol 22:2027–2039. doi:10.1093/molbev/msi193
Khaldi N, Seifuddin FT, Turner G et al (2010) SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47:736–741. doi:10.1016/j.fgb.2010.06.003
Khayatt BI, Overmars L, Siezen RJ, Francke C (2013) Classification of the adenylation and acyl-transferase activity of NRPS and PKS systems using ensembles of substrate specific hidden Markov models. PLoS ONE 8:e62136. doi:10.1371/journal.pone.0062136
Kim J, Yi G-S (2012) PKMiner: a database for exploring type II polyketide synthases. BMC Microbiol 12:169. doi:10.1186/1471-2180-12-169
Kwan DH, Leadlay PF (2010) Mutagenesis of a modular polyketide synthase enoylreductase domain reveals insights into catalysis and stereospecificity. ACS Chem Biol 5:829–838. doi:10.1021/cb100175a
Kwan DH, Sun Y, Schulz F et al (2008) Prediction and manipulation of the stereochemistry of enoylreduction in modular polyketide synthases. Chem Biol 15:1231–1240. doi:10.1016/j.chembiol.2008.09.012
Letunic I, Doerks T, Bork P (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37:D229–D232. doi:10.1093/nar/gkn808
Letzel A-C, Pidot SJ, Hertweck C (2013) A genomic approach to the cryptic secondary metabolome of the anaerobic world. Nat Prod Rep 30:392–428. doi:10.1039/c2np20103h
Li MHT, Ung PMU, Zajkowski J et al (2009) Automated genome mining for natural products. BMC Bioinformatics 10:185. doi:10.1186/1471-2105-10-185
Medema MH, Blin K, Cimermancic P et al (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. doi:10.1093/nar/gkr466
Metsä-Ketelä M, Halo L, Munukka E et al (2002) Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various streptomyces species. Appl Environ Microbiol 68:4472–4479
Minowa Y, Araki M, Kanehisa M (2007) Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J Mol Biol 368:1500–1517. doi:10.1016/j.jmb.2007.02.099
Nguyen T, Ishida K, Jenke-Kodama H et al (2008) Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol 26:225–233. doi:10.1038/nbt1379
Oliynyk M, Samborskyy M, Lester JB et al (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453. doi:10.1038/nbt1297
Penn K, Jenkins C, Nett M et al (2009) Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J 3:1193–1203. doi:10.1038/ismej.2009.58
Pickens LB, Kim W, Wang P et al (2009) Biochemical analysis of the biosynthetic pathway of an anticancer tetracycline SF2575. J Am Chem Soc 131:17677–17689. doi:10.1021/ja907852c
Piel J (2002) A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA 99:14002–14007. doi:10.1073/pnas.222481399
Piel J, Hui D, Wen G et al (2004) Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci USA 101:16222–16227. doi:10.1073/pnas.0405976101
Prieto C, García-Estrada C, Lorenzana D, Martín JF (2012) NRPSsp: non-ribosomal peptide synthase substrate predictor. Bioinformatics 28:426–427. doi:10.1093/bioinformatics/btr659
Rausch C, Hoof I, Weber T et al (2007) Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol 7:78. doi:10.1186/1471-2148-7-78
Reid R, Piagentini M, Rodriguez E et al (2003) A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72–79. doi:10.1021/bi0268706
Ridley CP, Lee HY, Khosla C (2008) Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci USA 105:4595–4600. doi:10.1073/pnas.0710107105
Röttig M, Medema MH, Blin K et al (2011) NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39:W362–W367. doi:10.1093/nar/gkr323
Schneiker S, Perlova O, Kaiser O et al (2007) Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25:1281–1289. doi:10.1038/nbt1354
Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6:493–505. doi:10.1016/S1074-5521(99)80082-9
Starcevic A, Zucko J, Simunkovic J et al (2008) ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res 36:6882–6892. doi:10.1093/nar/gkn685
Stevens DC, Conway KR, Pearce N et al (2013) Alternative sigma factor over-expression enables heterologous expression of a Type II polyketide biosynthetic pathway in Escherichia coli. PLoS ONE 8:e64858. doi:10.1371/journal.pone.0064858
Sun W, Peng C, Zhao Y, Li Z (2012) Functional gene-guided discovery of type II polyketides from culturable actinomycetes associated with soft coral Scleronephthya sp. PLoS ONE 7:e42847. doi:10.1371/journal.pone.0042847
Suwa M, Sugino H, Sasaoka A et al (2000) Identification of two polyketide synthase gene clusters on the linear plasmid pSLA2-L in Streptomyces rochei. Gene 246:123–131
Teta R, Gurgui M, Helfrich EJN et al (2010) Genome mining reveals trans-AT polyketide synthase directed antibiotic biosynthesis in the bacterial phylum bacteroidetes. ChemBioChem 11:2506–2512. doi:10.1002/cbic.201000542
Valenzano CR, You Y-O, Garg A et al (2010) Stereospecificity of the dehydratase domain of the erythromycin polyketide synthase. J Am Chem Soc 132:14697–14699. doi:10.1021/ja107344h
Vergnolle O, Hahn F, Baerga-Ortiz A et al (2011) Stereoselectivity of isolated dehydratase domains of the borrelidin polyketide synthase: implications for cis double bond formation. ChemBioChem 12:1011–1014. doi:10.1002/cbic.201100011
Volchegursky Y, Hu Z, Katz L, McDaniel R (2000) Biosynthesis of the anti-parasitic agent megalomicin: transformation of erythromycin to megalomicin in Saccharopolyspora erythraea. Mol Microbiol 37:752–762
Wang P, Kim W, Pickens LB et al (2012) Heterologous expression and manipulation of three tetracycline biosynthetic pathways. Angew Chem Int Ed Eng 51:11136–11140. doi:10.1002/anie.201205426
Weber T, Rausch C, Lopez P et al (2009) CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J Biotechnol 140:13–17
Wu J, Zaleski TJ, Valenzano C et al (2005) Polyketide double bond biosynthesis. Mechanistic analysis of the dehydratase-containing module 2 of the picromycin/methymycin polyketide synthase. J Am Chem Soc 127:17393–17404. doi:10.1021/ja055672+
Wyatt MA, Ahilan Y, Argyropoulos P et al (2013) Biosynthesis of ebelactone A: isotopic tracer, advanced precursor and genetic studies reveal a thioesterase-independent cyclization to give a polyketide β-lactone. J Antibiot. doi:10.1038/ja.2013.48
Wyatt MA, Lee J, Ahilan Y, Magarvey NA (2013) Bioinformatic evaluation of the secondary metabolism of antistaphylococcal environmental bacterial isolates. Can J Microbiol 59:465–471. doi:10.1139/cjm-2013-0016
Yadav G, Gokhale RS, Mohanty D (2003) Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol 328:335–363
Yadav G, Gokhale RS, Mohanty D (2009) Towards prediction of metabolic products of polyketide synthases: an in silico analysis. PLoS Comput Biol 5:e1000351. doi:10.1371/journal.pcbi.1000351
Zhang W, Ames BD, Tsai S-C, Tang Y (2006) Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl Environ Microbiol 72:2573–2580. doi:10.1128/AEM.72.4.2573- 2580.2006
Zhou H, Gao Z, Qiao K et al (2012) A fungal ketoreductase domain that displays substrate-dependent stereospecificity. Nat Chem Biol 8:331–333. doi:10.1038/nchembio.912
Ziemert N, Podell S, Penn K et al (2012) The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 7:e34064. doi:10.1371/journal.pone.0034064
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boddy, C.N. Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. J Ind Microbiol Biotechnol 41, 443–450 (2014). https://doi.org/10.1007/s10295-013-1368-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1368-1