Abstract
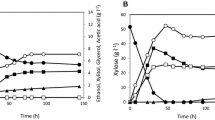
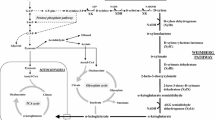
Robust microorganisms are necessary for economical bioethanol production. However, such organisms must be able to effectively ferment both hexose and pentose sugars present in lignocellulosic hydrolysate to ethanol. Wild type Saccharomyces cerevisiae can rapidly ferment hexose, but cannot ferment pentose sugars. Considerable efforts were made to genetically engineer S. cerevisiae to ferment xylose. Our genetically engineered S cerevisiae yeast, 424A(LNH-ST), expresses NADPH/NADH xylose reductase (XR) that prefer NADPH and NAD+-dependent xylitol dehydrogenase (XD) from Pichia stipitis, and overexpresses endogenous xylulokinase (XK). This strain is able to ferment glucose and xylose, as well as other hexose sugars, to ethanol. However, the preference for different cofactors by XR and XD might lead to redox imbalance, xylitol excretion, and thus might reduce ethanol yield and productivity. In the present study, genes responsible for the conversion of xylose to xylulose with different cofactor specificity (1) XR from N. crassa (NADPH-dependent) and C. parapsilosis (NADH-dependent), and (2) mutant XD from P. stipitis (containing three mutations D207A/I208R/F209S) were overexpressed in wild type yeast. To increase the NADPH pool, the fungal GAPDH enzyme from Kluyveromyces lactis was overexpressed in the 424A(LNH-ST) strain. Four pentose phosphate pathway (PPP) genes, TKL1, TAL1, RKI1 and RPE1 from S. cerevisiae, were also overexpressed in 424A(LNH-ST). Overexpression of GAPDH lowered xylitol production by more than 40%. However, other strains carrying different combinations of XR and XD, as well as new strains containing the overexpressed PPP genes, did not yield any significant improvement in xylose fermentation.




Similar content being viewed by others
References
Barnett JA (1976) The utilization of sugars by yeasts. Adv Carbohydr Chem Biochem 32:125–234
Lu C, Jeffries T (2007) Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol 73:6072–6077
Toivari MH, Aristidou A, Ruohonen L, Penttila M (2001) Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab Eng 3:236–249
Sedlak M, Ho NW (2004) Production of ethanol from cellulosic biomass hydrolysates using genetically engineered Saccharomyces yeast capable of cofermenting glucose and xylose. Appl Biochem Biotechnol 113–116:403–416
Sedlak M, Ho NW (2004) Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast 21:671–684
Verho R, Londesborough J, Penttila M, Richard P (2003) Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Appl Environ Microbiol 69:5892–5897
Richard P, Verho R, Putkonen M, Londesborough J, Penttila M (2003) Production of ethanol from L-arabinose by Saccharomyces cerevisiae containing a fungal L-arabinose pathway. FEMS Yeast Res 3:185–189
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386
Kotter P, Amore R, Hollenberg CP, Ciriacy M (1990) Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet 18:493–500
Ho NW, Chen Z, Brainard AP (1998) Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859
Ho NW, Chen Z, Brainard AP, Sedlak M (1999) Successful design and development of genetically engineered Saccharomyces yeasts for effective cofermentation of glucose and xylose from cellulosic biomass to fuel ethanol. Adv Biochem Eng Biotechnol 65:163–192
Kuyper M, Hartog MM, Toirkens MJ et al (2005) Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 5:399–409
Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT (2005) Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res 5:925–934
Jeppsson M, Traff K, Johansson B, Hahn-Hagerdal B, Gorwa-Grauslund MF (2003) Effect of enhanced xylose reductase activity on xylose consumption and product distribution in xylose-fermenting recombinant Saccharomyces cerevisiae. FEMS Yeast Res 3:167–175
Jin YS, Ni H, Laplaza JM, Jeffries TW (2003) Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate d-xylulokinase activity. Appl Environ Microbiol 69:495–503
Karhumaa K, Fromanger R, Hahn-Hagerdal B, Gorwa-Grauslund MF (2007) High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl Microbiol Biotechnol 73:1039–1046
Verduyn C, Van Kleef R, Frank J, Schreuder H, Van Dijken JP, Scheffers WA (1985) Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem J 226:669–677
Watanabe S, Saleh AA, Pack SP, Annaluru N, Kodaki T, Makino K (2007) Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+-dependent xylitol dehydrogenase. J Biotechnol 130:316–319
Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hagerdal B, Gorwa-Grauslund MF (2006) The expression of a Pichia stipitis xylose reductase mutant with higher K(M) for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 93:665–673
Matsushika A, Sawayama S (2008) Efficient bioethanol production from xylose by recombinant Saccharomyces cerevisiae requires high activity of xylose reductase and moderate xylulokinase activity. J Biosci Bioeng 106:306–309
Walfridsson M, Hallborn J, Penttila M, Keranen S, Hahn-Hagerdal B (1995) Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol 61:4184–4190
Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund MF (2005) Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22:359–368
Bera AK, Sedlak M, Khan A, Ho NW (2010) Establishment of L-arabinose fermentation in glucose/xylose co-fermenting recombinant Saccharomyces cerevisiae 424A(LNH-ST) by genetic engineering. Appl Microbiol Biotechnol 83:8003–8011
Casey E, Sedlak M, Ho NW, Mosier NS (2010) Effect of acetic acid and pH on the cofermentation of glucose and xylose to ethanol by a genetically engineered strain of Saccharomyces cerevisiae. FEMS Yeast Res 10:385–393
Taketo A (1988) DNA transfection of Escherichia coli by electroporation. Biochim Biophys Acta 949:318–324
Becker DM, Guarente L (1991) High-efficiency transformation of yeast by electroporation. Methods Enzymol 194:182–187
Chevallier MR, Aigle M (1979) Qualitative detection of penicillinase produced by yeast strains carrying chimeric yeast-coli plasmids. FEBS Lett 108:179–180
Krishnan MS, Ho NW, Tsao GT (1999) Fermentation kinetics of ethanol production from glucose and xylose by recombinant Saccharomyces 1400(pLNH33). Appl Biochem Biotechnol 77–79:373–388
Bruinenberg PM, van Dijken JP, Scheffers WA (1983) A theoretical analysis of NADPH production and consumption in yeasts. J Gen Microbiol 129:953–964
Petschacher B, Nidetzky B (2008) Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb Cell Fact 7:9
Lee JK, Koo BS, Kim SY (2003) Cloning and characterization of the xyl1 gene, encoding an NADH-preferring xylose reductase from Candida parapsilosis, and its functional expression in Candida tropicalis. Appl Environ Microbiol 69:6179–6188
Woodyer R, Simurdiak M, van der Donk WA, Zhao H (2005) Heterologous expression, purification, and characterization of a highly active xylose reductase from Neurospora crassa. Appl Environ Microbiol 71:1642–1647
Watanabe S, Kodaki T, Makino K (2005) Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J Biol Chem 280:10340–10349
Hou J, Shen Y, Li XP, Bao XM (2007) Effect of the reversal of coenzyme specificity by expression of mutated Pichia stipitis xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Lett Appl Microbiol 45:184–189
Annaluru N, Watanabe S, Pack SP, Saleh AA, Kodaki T, Makino K (2007) Thermostabilization of Pichia stipitis xylitol dehydrogenase by mutation of structural zinc-binding loop. J Biotechnol 129:717–722
Jeppsson M, Johansson B, Hahn-Hagerdal B, Gorwa-Grauslund MF (2002) Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. Appl Environ Microbiol 68:1604–1609
Metzger MH, Hollenberg CP (1994) Isolation and characterization of the Pichia stipitis transketolase gene and expression in a xylose-utilising Saccharomyces cerevisiae transformant. Appl Microbiol Biotechnol 42:319–325
Johansson B, Hahn-Hagerdal B (2002) The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res 2:277–282
Karhumaa K, Garcia Sanchez R, Hahn-Hagerdal B, Gorwa-Grauslund MF (2007) Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Fact 6:5
Dowell RD, Ryan O, Jansen A et al (2010) Genotype to phenotype: a complex problem. Science 328:469
Acknowledgments
The project was financially supported by the US Department of Energy Biomass Program, Contract G017059-16649.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is based on a presentation at the 32nd Symposium on Biotechnology for Fuels and Chemicals.
Rights and permissions
About this article
Cite this article
Bera, A.K., Ho, N.W.Y., Khan, A. et al. A genetic overhaul of Saccharomyces cerevisiae 424A(LNH-ST) to improve xylose fermentation. J Ind Microbiol Biotechnol 38, 617–626 (2011). https://doi.org/10.1007/s10295-010-0806-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0806-6