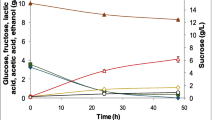
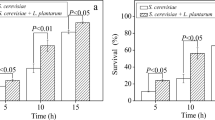
Abstract
Contamination of Lactobacillus sp. in the fermentation broth of bioethanol production decreases ethanol production efficiency. Although the addition of lactate to the broth can effectively inhibit the growth of Lactobacillus sp., it also greatly reduces the fermentation ability of Saccharomyces cerevisiae. To overcome this conflict, lactate-tolerant yeast strains were screened. Candida glabrata strain NFRI 3164 was found to exhibit both higher levels of lactate tolerance and fermentation ability. Co-cultivation of C. glabrata was performed with Lactobacillus brevis and Lb. fermentum, which were reported as major contaminating bacteria during bioethanol production, in culture medium containing 2% lactate. Under these culture conditions, the growth of Lactobacillus strains was greatly inhibited, but the ethanol production of C. glabrata was not significantly affected. Our data show the possibility of designing an effective fuel ethanol production process that eliminates contamination by Lactobacillus strains through the combined use of lactate addition and C. glabrata.



Similar content being viewed by others
References
Antoni D, Zverlov VV, Schwarz WH (2007) Biofuels from microbes. Appl Microbiol Biotechnol 77:23–35
Aquarone E (1960) Penicillin and tetracycline as contamination control agents in alcoholic fermentation of sugar cane molasses. Appl Microbiol 8:263–268
Barnett JA, Payne RW, Yaroww D (2000) Yeasts: characteristics and identification. Cambridge University Press, Cambridge
Bayrock DP, Thomas KC, Ingledew WM (2003) Control of Lactobacillus contaminants in continuous fuel ethanol fermentations by constant or pulsed addition of penicillin G. Appl Microbiol Biotechnol 62:498–502
Chang IS, Kim BH, Shin PK (1997) Use of sulfite and hydrogen peroxide to control bacterial contamination in ethanol fermentation. Appl Environ Microbiol 63:1–6
Dawson L, Boopathy R (2007) Use of post-harvest sugarcane residue for ethanol production. Bioresour Technol 98:1695–1699
Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wesolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL (2004) Genome evolution in yeasts. Nature 430:35–44
Gray KA, Zhao L, Emptage M (2006) Bioethanol. Curr Opin Chem Biol 10:141–146
Gregori C, Schuller C, Roetzer A, Schwarzmuller T, Ammerer G, Kuchler K (2007) The high-osmolarity glycerol response pathway in the human fungal pathogen Candida glabrata strain ATCC 2001 lacks a signaling branch that operates in baker’s yeast. Eukaryot Cell 6:1635–1645
Herrero E (2005) Evolutionary relationships between Saccharomyces cerevisiae and other fungal species as determined from genome comparisons. Rev Iberoam Micol 22:217–222
Hynes SH, Kjarsgaard DM, Thomas KC, Ingledew WM (1997) Use of virginiamycin to control the growth of lactic acid bacteria during alcohol fermentation. J Ind Microbiol Biotechnol 18:284–291
Kadam KL, McMillan JD (2003) Availability of corn stover as a sustainable feedstock for bioethanol production. Bioresour Technol 88:17–25
Kim S, Dale B (2005) Life cycle assessment of various cropping systems utilized for producing biofuels: bioethanol and biodiesel. Biomass Bioenerg 29:426–439
Kreger-van Rij NJW, Jack WF (1998) The yeasts: a taxonomic study. Elsevier Science Ltd, Amsterdam
Kuriyama H, Seiko Y, Murakami T, Kobayashi H, Sonoda Y, Silva H (1985) Selection of yeast strains for effective ethanol fermentation. Rep Ferment Res Inst 64:43–50
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Levine AS, Fellers CR (1940) Action of acetic acid on food spoilage microorganisms. J Bacteriol 39:499–515
Maiorella B, Blanch HW, Wilke CR (1983) By-product inhibition effects on ethanolic fermentation by Saccharomyces cerevisiae. Biotechnol Bioeng 25:103–131
Makanjuola DB, Tymon A, Springham DG (1992) Some effects of lactic acid bacteria on laboratory-scale yeast fermentations. Enzyme Microb Technol 14:350–357
Merico A, Sulo P, Piskur J, Compagno C (2007) Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J 274:976–989
Nakagawa Y, Mizuguchi I (2005) The nucleotide sequence determination of catalases of three medically important yeasts using newly designed degenerated primers. Nihon Ishinkin Gakkai Zasshi/Jpn J Med Mycol 46:35–42
Narendranath NV, Thomas KC, Ingledew WM (2001) Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in a minimal medium. J Ind Microbiol Biotechnol 26:171–177
Narendranath NV, Thomas KC, Ingledew WM (2000) Urea hydrogen peroxide reduces the numbers of lactobacilli, nourishes yeast, and leaves no residues in the ethanol fermentation. Appl Environ Microbiol 66:4187–4192
Nikkuni S (1990) Annual report on exploration and introduction of microbial genetic resources. Annu Rep Natl Inst Agrobiol Resour 2:59–71
Nishida O, Kuwazaki S, Suzuki C, Shima J (2004) Superior molasses assimilation, stress tolerance, and trehalose accumulation of baker’s yeast isolated from dried sweet potatoes (hoshi-imo). Biosci Biotechnol Biochem 68:1442–1448
Nishino N, Hattori H, Kishida Y (2007) Alcoholic fermentation and its prevention by Lactobacillus buchneri in whole crop rice silage. Lett Appl Microbiol 44:538–543
Oliva-Neto PD, Yokoya F (1998) Effect of 3,4,4′-trichlorocarbanilide on growth of lactic acid bacteria contaminants in alcoholic fermentation. Bioresour Technol 63:17–21
Pikur J, Langkjr RB (2004) Yeast genome sequencing: the power of comparative genomics. Mol Microbiol 53:381–389
Rudolf A, Baudel H, Zacchi G, Hahn-Hagerdal B, Liden G (2008) Simultaneous saccharification and fermentation of steam-pretreated bagasse using Saccharomyces cerevisiae TMB3400 and Pichia stipitis CBS6054. Biotechnol Bioeng 99(4):783–790
Schell DJ, Dowe N, Ibsen KN, Riley CJ, Ruth MF, Lumpkin RE (2007) Contaminant occurrence, identification and control in a pilot-scale corn fiber to ethanol conversion process. Bioresour Technol 98:2942–2948
Schell DJ, Riley CJ, Dowe N, Farmer J, Ibsen KN, Ruth MF, Toon ST, Lumpkin RE (2004) A bioethanol process development unit: initial operating experiences and results with a corn fiber feedstock. Bioresour Technol 91:179–188
Seiko Y, Murakami T, Kuriyama H, Sonoda Y (1985) Selection of yeast strains having tolerance to inhibitive conditions. Rep Ferment Res Inst 63:55–63
Sherman F, Fink G, Lawrence C (eds) (1979) Methods in yeast genetics. Cold Spring Harbor Laboratory, New York
Simpson KL, Pettersson B, Priest FG (2001) Characterization of lactobacilli from Scotch malt whisky distilleries and description of Lactobacillus ferintoshensis sp. nov., a new species isolated from malt whisky fermentations. Microbiology 147:1007–1016
Skinner KA, Leathers TD (2004) Bacterial contaminants of fuel ethanol production. J Ind Microbiol Biotechnol 31:401–408
Solomon BD, Barnes JR, Halvorsen KE (2007) Grain and cellulosic ethanol: history, economics, and energy policy. Biomass Bioenerg 31:416–425
Thomas KC, Hynes SH, Ingledew WM (2001) Effect of lactobacilli on yeast growth, viability and batch and semi-continuous alcoholic fermentation of corn mash. J Appl Microbiol 90:819–828
Torney F, Moeller L, Scarpa A, Wang K (2007) Genetic engineering approaches to improve bioethanol production from maize. Curr Opin Biotechnol 18:193–199
Vinderola CG, Reinheimer JA (2003) Lactic acid starter and probiotic bacteria: a comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res Int 36:895–904
Visser W, Scheffers WA, Batenburg-van der Vegte WH, van Dijken JP (1990) Oxygen requirements of yeasts. Appl Environ Microbiol 56:3785–3792
von Blottnitz H, Curran MA (2007) A review of assessments conducted on bio-ethanol as a transportation fuel from a net energy, greenhouse gas, and environmental life cycle perspective. J Cean Prod 15:607–619
Wheals AE, Basso LC, Alves DM, Amorim HV (1999) Fuel ethanol after 25 years. Trends Biotechnol 17:482–487
William RG, Carl AW (1986) Effects of sodium meta bisulfite on diffusion fermentation of fodder beets for fuel ethanol production. Biotechnol Bioeng 30:909–916
Acknowledgments
This work was supported by the project for the development of biomass utilization technologies for revitalizing rural areas from the Ministry of Agriculture, Forestry, and Fisheries (MAFF) of Japan. We thank S. Mizukami-Murata and Y. Sakayori (National Food Research Institute) for their technical assistance in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Watanabe, I., Nakamura, T. & Shima, J. A strategy to prevent the occurrence of Lactobacillus strains using lactate-tolerant yeast Candida glabrata in bioethanol production. J Ind Microbiol Biotechnol 35, 1117–1122 (2008). https://doi.org/10.1007/s10295-008-0390-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0390-1