Abstract
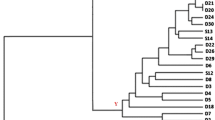
This study reports on monitoring Oenococcus oeni intraspecific diversity evolution during winemaking. Three different wines were monitored. The proportion of O. oeni species was determined by species-specific PCR and O. oeni strains were distinguished by multiplex PCR-RAPD. Each strain was tested by PCR for 16 significant markers revealed by a previous genetic comparison between a strong oenological potential strain and one with poor oenological potential. Population levels and diversity changed according to winemaking stages, oenological practices and the chemical properties of the wine. In all situations, O. oeni was the best-adapted species. Within the O. oeni group, intraspecific strain diversity decreased and the malolactic fermentation was the result of the most resistant strains with the highest number of markers.



Similar content being viewed by others
References
Banjerdkij P, Vattanaviboon P, Mongkolsuk S (2004) Cadmium-induced adaptive resistance and cross-resistance to zinc in Xanthomonas campestris. Curr Microbiol 47:260–262
Brynhildsen L, Lundgren BV, Allard B, Rosswall T (1988) Effects of glucose concentrations on cadmium, copper, mercury, and zinc toxicity to a Klebsiella sp. Appl Environ Microbiol 54:1689–1693
Campos FM, Couto JA, Hogg TA (2003) Influence of phenolic acids on growth and inactivation of Oenococcus oeni and Lactobacillus hilgardii. J Appl Microbiol 94:167–174; Microbiol Lett 246:111–117
Cocconcelli PS, Porro D, Galandini S, Sentini L (1995) Development of RAPD protocol for typing of strains of lactic acid bacteria and enterococci. Lett Appl Microbiol 21:376–379
Delaherche A (2006) Adaptation d’Oenococcus oeni à l’environnement œnologique approches génomiques comparées, transcriptomiques et protéomiques. Ph D Thesis. Université Bordeaux 2, France
Delaherche A, Bon E, Gindreau E, de Daruvar A, Lonvaud-Funel A (2007a) The use of comparative genomic analysis to identify distinct genetic markers of Oenococcus oeni during adaptation in wine. Appl Environ Microbiol (in press)
Delaherche A, Bon E, Dupré , Lucas M, Arveiller B, De Daruvar A, Lonvaud-Funel A (2007b) Intraspecific diversity of Oenococcus oeni strains determined by sequence analysis of target genes. Appl Microbiol Biotechnol 73:394–403
Divol B, Tonon T, Morichon S, Gindreau E, Lonvaud-Funel A (2003) Molecular characterization of Oenococcus oeni genes encoding proteins involved in arginine transport. J Appl Microbiol 94:738–746
Gindreau E, Joyeux A, De Revel G, Claisse O, Lonvaud-Funel A (1997) Evaluation de l’établissement de levains malolactiques au sein de la microflore bactérienne indigène. J Int Sci Vigne Vin 31:197–202
Guzzo J, Jobin MP, Delmas F, Fortier LC, Garmyn D, Tourdot-Marechal R, Lee B, Divies C (2000) Regulation of stress response in Oenococcus oeni as a function of environmental changes and growth phase. Int J Food Microbiol 55:27–31
Lemaresquier H (1987) Inter-relationships between strains of Saccharomyces cerevisiae from the Champagne area and lactic acid bacteria. Lett Appl Microbiol 4:91–94
Reguant C, Bordons A (2003) Typication of Oenococcus oeni strains by multiplex RAPD-PCR and study of population dynamics during malolactic fermentation. J Appl Microbiol 95:244–353
Renouf V, Claisse O, Lonvaud-Funel A (2005a) Understanding the microbial ecosystem on the grape berry surface through numeration and identification of yeast and bacteria. Aust J Grape Wine Res 11:316–327
Renouf V, Walling E, Coulon J, Lonvaud-Funel A (2006a) Conseils pratiques pour la mise en place d’un suivi microbiologique des vins. Rev Oenol 119:39–42
Renouf V, Claisse O, Miot-Sertier C, Lonvaud-Funel A (2006b) Lactic acid bacteria evolution during winemaking: use of the rpoB gene as a target for PCR-DGGE analysis. Food Microbiol 23:136–145
Renouf V, Claisse O, Lonvaud-Funel A (2007) Inventory and monitoring of wine microbial consortia. Appl Microbiol Biotechnol 75:149–164
van der Heide T, Boolman B (2000) Osmoregulated ABC transport system of Lactococcus lactis senses water stress via changes in the physical state of membrane. Biochemistry 97:7102–7106
Wibowo D, Eschenbruch R, Davis CR, Fleet GH, Lee TH (1985) Occurrence and growth of lactic acid bacteria in wine. A review. Am J Enol Vitic 36:302–313
Theil EC (2007) Coordinating responses to iron and oxygen stress with DNA and mRNA promoters: the ferritin story. Biometals (in press)
Zapparolli G, Torriani S, Pesente P, Dellaglio F (1998) Design and evaluation of malolactic enzyme gene targeted primers for rapid identification and detection of Oenoccocus oeni in wine. Lett Appl Microbiol 27:243–246
Zeller T, Klug G (2006) Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften 93:259–266
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Renouf, V., Delaherche, A., Claisse, O. et al. Correlation between indigenous Oenococcus oeni strain resistance and the presence of genetic markers. J Ind Microbiol Biotechnol 35, 27–33 (2008). https://doi.org/10.1007/s10295-007-0262-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-007-0262-0