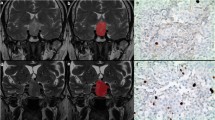
Abstract
Objectives
To preoperatively predict the high expression of Ki67 and positive pituitary transcription factor 1 (PIT-1) simultaneously in pituitary adenoma (PA) using three different radiomics models.
Methods
A total of 247 patients with PA (training set: n = 198; test set: n = 49) were included in this retrospective study. The imaging features were extracted from preoperative contrast-enhanced T1WI (T1CE), T1-weighted imaging (T1WI), and T2-weighted imaging (T2WI). Feature selection was performed using Spearman’s rank correlation coefficient and least absolute shrinkage and selection operator (LASSO). The classic machine learning (CML), deep learning (DL), and deep learning radiomics (DLR) models were constructed using logistic regression (LR), support vector machine (SVM), and multi-layer perceptron (MLP) algorithms. The area under the receiver operating characteristic (ROC) curve (AUC), sensitivity, specificity, accuracy, negative predictive value (NPV) and positive predictive value (PPV) were calculated for the training and test sets. In addition, combined with clinical characteristics, the best CML and the best DL models (SVM classifier), the DL radiomics nomogram (DLRN) was constructed to aid clinical decision-making.
Results
Seven CML features, 96 DL features, and 107 DLR features were selected to construct CML, DL and DLR models. Compared to CML and DL model, the DLR model had the best performance. The AUC, sensitivity, specificity, accuracy, NPV and PPV were 0.827, 0.792, 0.800, 0.796, 0.800 and 0.792 in the test set, respectively.
Conclusions
Compared with CML and DL models, the DLR model shows the best performance in predicting the Ki67 and PIT-1 expression in PAs simultaneously.









Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- CML:
-
Classic machine learning
- DCA:
-
Decision curve analysis
- DICOM:
-
Digital imaging and communications in medicine
- DL:
-
Deep learning
- DLR:
-
Deep learning radiomics
- DLRN:
-
DL radiomics nomogram
- GH:
-
Growth hormone
- Grad-CAM:
-
Gradient-weighted class activation mapping
- LASSO:
-
Least absolute shrinkage and selection operator
- LR:
-
Logistic regression
- MLP:
-
Multi-layer perceptron
- MSE:
-
Mean Standard Error
- NPV:
-
Negative predictive value
- PA:
-
Pituitary adenoma
- PIT-1:
-
Positive pituitary transcription factor 1
- PPV:
-
Positive predictive value
- ROI:
-
Region of interest
- ROC:
-
Receiver operating characteristic
- SVM:
-
Support vector machine
- T1CE:
-
Contrast-enhanced T1-weighted imaging
- T1WI:
-
T1-weighted imaging
- T2WI:
-
T2-weighted imaging
5 References
S. Melmed, U.B. Kaiser, M.B. Lopes, J. Bertherat, L.V. Syro, G. Raverot, M. Reincke, G. Johannsson, A. Beckers, M. Fleseriu, A. Giustina, J.A.H. Wass, K.K.Y. Ho, Clinical Biology of the Pituitary Adenoma, Endocr. Rev. 43(6) (2022) 1003–1037, https://doi.org/10.1210/endrev/bnac010
M.W. Møller, M.S. Andersen, D. Glintborg, C.B. Pedersen, B. Halle, B.W. Kristensen, F.R. Poulsen, [Pituitary adenoma], Ugeskr. Laeger 181(20) (2019),
C.S. Graffeo, K.J. Yagnik, L.P. Carlstrom, N. Lakomkin, I. Bancos, C. Davidge-Pitts, D. Erickson, G. Choby, B.E. Pollock, A.M. Chamberlain, J.J. Van Gompel, Pituitary Adenoma Incidence, Management Trends, and Long-term Outcomes: A 30-Year Population-Based Analysis, Mayo Clinic Proceedings 97(10) (2022) 1861–1871, https://doi.org/10.1016/j.mayocp.2022.03.017
A.F. Daly, A. Beckers, The Epidemiology of Pituitary Adenomas, Endocrinol. Metab. Clin. North Am. 49(3) (2020) 347–355, https://doi.org/10.1016/j.ecl.2020.04.002
G. Raverot, M.D. Ilie, H. Lasolle, V. Amodru, J. Trouillas, F. Castinetti, T. Brue, Aggressive pituitary tumours and pituitary carcinomas, Nat. Rev. Endocrinol. 17(11) (2021) 671–684, https://doi.org/10.1038/s41574-021-00550-w
I.F. Burcea, V.N. Năstase, C. Poiană, Pituitary transcription factors in the immunohistochemical and molecular diagnosis of pituitary tumours - a systematic review, Endokrynol. Pol. 72(1) (2021) 53–63, https://doi.org/10.5603/EP.a2020.0090
K. Asmaro, M. Zhang, A.J. Rodrigues, A. Mohyeldin, V. Vigo, K. Nernekli, H. Vogel, D.E. Born, L. Katznelson, J.C. Fernandez-Miranda, Cytodifferentiation of pituitary tumors influences pathogenesis and cavernous sinus invasion, J. Neurosurg. 139(5) (2023) 1216–1224, https://doi.org/10.3171/2023.3.Jns221949
M. Luo, R. Tang, H. Wang, Tumor immune microenvironment in pituitary neuroendocrine tumors (PitNETs): increased M2 macrophage infiltration and PD-L1 expression in PIT1-lineage subset, J. Neurooncol. 163(3) (2023) 663–674, https://doi.org/10.1007/s11060-023-04382-8
X.Y. Wan, J. Chen, J.W. Wang, Y.C. Liu, K. Shu, T. Lei, Overview of the 2022 WHO Classification of Pituitary Adenomas/Pituitary Neuroendocrine Tumors: Clinical Practices, Controversies, and Perspectives, Curr Med Sci 42(6) (2022) 1111–1118, https://doi.org/10.1007/s11596-022-2673-6
X.Y. Liu, M.G. Wei, J. Liang, H.H. Xu, J.J. Wang, J. Wang, X.P. Yang, F.F. Lv, K.Q. Wang, J.H. Duan, Y. Tu, S. Zhang, C. Chen, X.H. Li, Injury-preconditioning secretome of umbilical cord mesenchymal stem cells amplified the neurogenesis and cognitive recovery after severe traumatic brain injury in rats, Journal of Neurochemistry 153(2) (2020) 230–251, https://doi.org/10.1111/jnc.14859
L. Yuhan, W. Zhiqun, T. Jihui, P. Renlong, Ki-67 labeling index and Knosp classification of pituitary adenomas, Br. J. Neurosurg. (2021) 1–5, https://doi.org/10.1080/02688697.2021.1884186
L. Mastronardi, A. Guiducci, C. Spera, F. Puzzilli, F. Liberati, G. Maira, Ki-67 labelling index and invasiveness among anterior pituitary adenomas: analysis of 103 cases using the MIB-1 monoclonal antibody, J. Clin. Pathol. 52(2) (1999) 107–111, https://doi.org/10.1136/jcp.52.2.107
C. Xue, S. Liu, J. Deng, X. Liu, S. Li, P. Zhang, J. Zhou, Apparent Diffusion Coefficient Histogram Analysis for the Preoperative Evaluation of Ki-67 Expression in Pituitary Macroadenoma, Clin Neuroradiol 32(1) (2022) 269–276, https://doi.org/10.1007/s00062-021-01134-x
A. Conficoni, P. Feraco, D. Mazzatenta, M. Zoli, S. Asioli, C. Zenesini, V.P. Fabbri, M. Cellerini, A. Bacci, Biomarkers of pituitary macroadenomas aggressive behaviour: a conventional MRI and DWI 3T study, Br. J. Radiol. 93(1113) (2020) 20200321, https://doi.org/10.1259/bjr.20200321
X.-j. Shu, H. Chang, Q. Wang, W.-g. Chen, K. Zhao, B.-y. Li, G.-c. Sun, S.-b. Chen, B.-n. Xu, Deep Learning model-based approach for preoperative prediction of Ki67 labeling index status in a noninvasive way using magnetic resonance images: A single-center study, Clinical Neurology and Neurosurgery 219 (2022), https://doi.org/10.1016/j.clineuro.2022.107301
L. Ugga, R. Cuocolo, D. Solari, E. Guadagno, A. D’Amico, T. Somma, P. Cappabianca, M.L. Del Basso de Caro, L.M. Cavallo, A. Brunetti, Prediction of high proliferative index in pituitary macroadenomas using MRI-based radiomics and machine learning, Neuroradiology 61(12) (2019) 1365–1373, https://doi.org/10.1007/s00234-019-02266-1
A. Peng, H. Dai, H. Duan, Y. Chen, J. Huang, L. Zhou, L. Chen, A machine learning model to precisely immunohistochemically classify pituitary adenoma subtypes with radiomics based on preoperative magnetic resonance imaging, European Journal of Radiology 125 (2020), https://doi.org/10.1016/j.ejrad.2020.108892
Y. Chen, F. Cai, J. Cao, F. Gao, Y. Lv, Y. Tang, A. Zhang, W. Yan, Y. Wang, X. Hu, S. Chen, X. Dong, J. Zhang, Q. Wu, Analysis of Related Factors of Tumor Recurrence or Progression After Transnasal Sphenoidal Surgical Treatment of Large and Giant Pituitary Adenomas and Establish a Nomogram to Predict Tumor Prognosis, Front Endocrinol (Lausanne) 12 (2021) 793337, https://doi.org/10.3389/fendo.2021.793337
X.J. Shu, H. Chang, Q. Wang, W.G. Chen, K. Zhao, B.Y. Li, G.C. Sun, S.B. Chen, B.N. Xu, Deep Learning model-based approach for preoperative prediction of Ki67 labeling index status in a noninvasive way using magnetic resonance images: A single-center study, Clin. Neurol. Neurosurg. 219 (2022) 107301, https://doi.org/10.1016/j.clineuro.2022.107301
S. Aydin, N. Comunoglu, M.L. Ahmedov, O.P. Korkmaz, B. Oz, P. Kadioglu, N. Gazioglu, N. Tanriover, Clinicopathologic Characteristics and Surgical Treatment of Plurihormonal Pituitary Adenomas, World Neurosurg 130 (2019) e765-e774, https://doi.org/10.1016/j.wneu.2019.06.217
X. Li, P.S. Morgan, J. Ashburner, J. Smith, C. Rorden, The first step for neuroimaging data analysis: DICOM to NIfTI conversion, J Neurosci Methods 264 (2016) 47–56, https://doi.org/10.1016/j.jneumeth.2016.03.001
A. Myronenko, M.M.R. Siddiquee, D. Yang, Y. He, D. Xu, Automated head and neck tumor segmentation from 3D PET/CT, HECKTOR@MICCAI, 2022.
L.R. Dice, MEASURES OF THE AMOUNT OF ECOLOGIC ASSOCIATION BETWEEN SPECIES, Ecology 26(3) (1945) 297–302, https://doi.org/10.2307/1932409
J. Fan, M. Chen, J. Luo, S. Yang, J. Shi, Q. Yao, X. Zhang, S. Du, H. Qu, Y. Cheng, S. Ma, M. Zhang, X. Xu, Q. Wang, S. Zhan, The prediction of asymptomatic carotid atherosclerosis with electronic health records: a comparative study of six machine learning models, BMC Med. Inform. Decis. Mak. 21(1) (2021) 115, https://doi.org/10.1186/s12911-021-01480-3
P. Schober, T.R. Vetter, Logistic Regression in Medical Research, Anesth Analg 132(2) (2021) 365–366, https://doi.org/10.1213/ane.0000000000005247
J.J. Raposo-Neto, E. Kowalski-Neto, W.B. Luiz, E.A. Fonseca, A. Cedro, M.N. Singh, F.L. Martin, P.F. Vassallo, L.C.G. Campos, V.G. Barauna, Near-Infrared Spectroscopy with Supervised Machine Learning as a Screening Tool for Neutropenia, J Pers Med 14(1) (2023), https://doi.org/10.3390/jpm14010009
H.Y. Wu, C.A. Gong, S.P. Lin, K.Y. Chang, M.Y. Tsou, C.K. Ting, Predicting postoperative vomiting among orthopedic patients receiving patient-controlled epidural analgesia using SVM and LR, Sci Rep 6 (2016) 27041, https://doi.org/10.1038/srep27041
N.A. Almansour, H.F. Syed, N.R. Khayat, R.K. Altheeb, R.E. Juri, J. Alhiyafi, S. Alrashed, S.O. Olatunji, Neural network and support vector machine for the prediction of chronic kidney disease: A comparative study, Comput. Biol. Med. 109 (2019) 101–111, https://doi.org/10.1016/j.compbiomed.2019.04.017
K. Nagawa, M. Suzuki, Y. Yamamoto, K. Inoue, E. Kozawa, T. Mimura, K. Nakamura, M. Nagata, M. Niitsu, Texture analysis of muscle MRI: machine learning-based classifications in idiopathic inflammatory myopathies, Sci Rep 11(1) (2021) 9821, https://doi.org/10.1038/s41598-021-89311-3
C.Y. Hsu, P.Y. Liu, S.H. Liu, Y. Kwon, C.J. Lavie, G.M. Lin, Machine Learning for Electrocardiographic Features to Identify Left Atrial Enlargement in Young Adults: CHIEF Heart Study, Front Cardiovasc Med 9 (2022) 840585, https://doi.org/10.3389/fcvm.2022.840585
R.R. Selvaraju, M. Cogswell, A. Das, R. Vedantam, D. Parikh, D. Batra, Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization, International Journal of Computer Vision 128(2) (2020) 336–359, https://doi.org/10.1007/s11263-019-01228-7
A.J. Vickers, E.B. Elkin, Decision curve analysis: a novel method for evaluating prediction models, Med Decis Making 26(6) (2006) 565–574, https://doi.org/10.1177/0272989x06295361
D.M. Wei, W.J. Chen, R.M. Meng, N. Zhao, X.Y. Zhang, D.Y. Liao, G. Chen, Augmented expression of Ki-67 is correlated with clinicopathological characteristics and prognosis for lung cancer patients: an up-dated systematic review and meta-analysis with 108 studies and 14,732 patients, Respir. Res. 19(1) (2018) 150, https://doi.org/10.1186/s12931-018-0843-7
G. Zada, N. Lin, E.R. Laws, Jr., Patterns of extrasellar extension in growth hormone-secreting and nonfunctional pituitary macroadenomas, Neurosurg. Focus 29(4) (2010) E4, https://doi.org/10.3171/2010.7.Focus10155
X. Cai, J. Zhu, J. Yang, C. Tang, F. Yuan, Z. Cong, C. Ma, A Nomogram for Preoperatively Predicting the Ki-67 Index of a Pituitary Tumor: A Retrospective Cohort Study, Frontiers in Oncology 11 (2021), https://doi.org/10.3389/fonc.2021.687333
H. Li, Z. Liu, F. Li, F. Shi, Y. Xia, Q. Zhou, Q. Zeng, Preoperatively Predicting Ki67 Expression in Pituitary Adenomas Using Deep Segmentation Network and Radiomics Analysis Based on Multiparameter MRI, Academic Radiology (2023), https://doi.org/10.1016/j.acra.2023.05.023
Z. Li, Y. Wang, J. Yu, Y. Guo, W. Cao, Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma, Scientific Reports 7(1) (2017), https://doi.org/10.1038/s41598-017-05848-2
X. Dong, J. Yang, B. Zhang, Y. Li, G. Wang, J. Chen, Y. Wei, H. Zhang, Q. Chen, S. Jin, L. Wang, H. He, M. Gan, W. Ji, Deep Learning Radiomics Model of Dynamic Contrast-Enhanced MRI for Evaluating Vessels Encapsulating Tumor Clusters and Prognosis in Hepatocellular Carcinoma, Journal of Magnetic Resonance Imaging 59(1) (2023) 108–119, https://doi.org/10.1002/jmri.28745
M.K.H. Khan, W. Guo, J. Liu, F. Dong, Z. Li, T.A. Patterson, H. Hong, Machine learning and deep learning for brain tumor MRI image segmentation, Experimental Biology and Medicine (2023), https://doi.org/10.1177/15353702231214259
X. Kong, Y. Mao, F. Xi, Y. Li, Y. Luo, J. Ma, Development of a nomogram based on radiomics and semantic features for predicting chromosome 7 gain/chromosome 10 loss in IDH wild-type histologically low-grade gliomas, Frontiers in Oncology 13 (2023), https://doi.org/10.3389/fonc.2023.1196614
J. Li, T. Zhang, J. Ma, N. Zhang, Z. Zhang, Z. Ye, Machine-learning-based contrast-enhanced computed tomography radiomic analysis for categorization of ovarian tumors, Front Oncol 12 (2022) 934735, https://doi.org/10.3389/fonc.2022.934735
K. Yun, T. He, S. Zhen, M. Quan, X. Yang, D. Man, S. Zhang, W. Wang, X. Han, Development and validation of explainable machine-learning models for carotid atherosclerosis early screening, J. Transl. Med. 21(1) (2023) 353, https://doi.org/10.1186/s12967-023-04093-8
J. Wang, H. Chen, H. Wang, W. Liu, D. Peng, Q. Zhao, M. Xiao, A Risk Prediction Model for Physical Restraints Among Older Chinese Adults in Long-term Care Facilities: Machine Learning Study, J. Med. Internet Res. 25 (2023) e43815, https://doi.org/10.2196/43815
A. Peng, H. Dai, H. Duan, Y. Chen, J. Huang, L. Zhou, L. Chen, A machine learning model to precisely immunohistochemically classify pituitary adenoma subtypes with radiomics based on preoperative magnetic resonance imaging, Eur. J. Radiol. 125 (2020) 108892, https://doi.org/10.1016/j.ejrad.2020.108892
Y. Zheng, D. Zhou, H. Liu, M. Wen, CT-based radiomics analysis of different machine learning models for differentiating benign and malignant parotid tumors, Eur. Radiol. 32(10) (2022) 6953–6964, https://doi.org/10.1007/s00330-022-08830-3
Y. Fan, S. Jiang, M. Hua, S. Feng, M. Feng, R. Wang, Machine Learning-Based Radiomics Predicts Radiotherapeutic Response in Patients With Acromegaly, Frontiers in Endocrinology 10 (2019), https://doi.org/10.3389/fendo.2019.00588
R. Cuocolo, L. Ugga, D. Solari, S. Corvino, A. D’Amico, D. Russo, P. Cappabianca, L.M. Cavallo, A. Elefante, Prediction of pituitary adenoma surgical consistency: radiomic data mining and machine learning on T2-weighted MRI, Neuroradiology 62(12) (2020) 1649–1656, https://doi.org/10.1007/s00234-020-02502-z
H. Wang, W. Zhang, S. Li, Y. Fan, M. Feng, R. Wang, Development and Evaluation of Deep Learning-based Automated Segmentation of Pituitary Adenoma in Clinical Task, The Journal of Clinical Endocrinology & Metabolism 106(9) (2021) 2535–2546, https://doi.org/10.1210/clinem/dgab371
G. Raverot, P. Burman, A. McCormack, A. Heaney, S. Petersenn, V. Popovic, J. Trouillas, O.M. Dekkers, European Society of Endocrinology Clinical Practice Guidelines for the management of aggressive pituitary tumours and carcinomas, European Journal of Endocrinology 178(1) (2018) G1-G24, https://doi.org/10.1530/eje-17-0796
S.K. Cheok, J.D. Carmichael, G. Zada, Management of growth hormone–secreting pituitary adenomas causing acromegaly: a practical review of surgical and multimodal management strategies for neurosurgeons, Journal of Neurosurgery (2023) 1–10, https://doi.org/10.3171/2023.8.Jns221975
A. Ishida, H. Shiramizu, H. Yoshimoto, M. Kato, N. Inoshita, N. Miki, M. Ono, S. Yamada, Resection of the Cavernous Sinus Medial Wall Improves Remission Rate in Functioning Pituitary Tumors: Retrospective Analysis of 248 Consecutive Cases, Neurosurgery 91(5) (2022) 775–781, https://doi.org/10.1227/neu.0000000000002109
F. Salehi, A. Agur, B.W. Scheithauer, K. Kovacs, R.V. Lloyd, M. Cusimano, Ki-67 in Pituitary Neoplasms, Neurosurgery 65(3) (2009) 429–437, https://doi.org/10.1227/01.Neu.0000349930.66434.82
J.P. Andrews, R.S. Joshi, M.P. Pereira, T. Oh, A.F. Haddad, K.M. Pereira, R.C. Osorio, K.C. Donohue, Z. Peeran, S. Sudhir, S. Jain, A. Beniwal, A.S. Chopra, N.S. Sandhu, T. Tihan, L. Blevins, M.K. Aghi, Plurihormonal PIT-1–Positive Pituitary Adenomas: A Systematic Review and Single-Center Series, World Neurosurgery 151 (2021) e185-e191, https://doi.org/10.1016/j.wneu.2021.04.003
Acknowledgements
We thank Fei Zheng, M.M., Department of Radiology, Peking University People’s Hospital, for her assistance in methods.
Funding
This study has received funding by the Beijing Hospitals Authority Clinical Medicine Development of Special (XMLX202108) and the collaborative innovative major special project supported by Beijing Municipal Science & Technology Commission (No. Z191100006619088).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
This retrospective study was approved by the Ethics Committee of Beijing Tiantan Hospital, Capital Medical University.
Consent to Participate
Written informed consent was waived by the Institutional Review Board.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, F., Zang, Y., Feng, L. et al. Concomitant Prediction of the Ki67 and PIT-1 Expression in Pituitary Adenoma Using Different Radiomics Models. J Digit Imaging. Inform. med. (2024). https://doi.org/10.1007/s10278-024-01121-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10278-024-01121-x