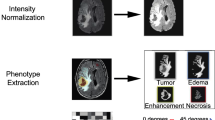
Abstract
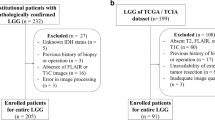
We aimed to study whether the Bruton’s tyrosine kinase (BTK) expression is correlated with the prognosis of patients with high-grade gliomas (HGGs) and predict its expression level prior to surgery, by constructing radiomic models. Clinical and gene expression data of 310 patients from The Cancer Genome Atlas (TCGA) were included for gene-based prognostic analysis. Among them, contrast-enhanced T1-weighted imaging (T1WI + C) from The Cancer Imaging Archive (TCIA) with genomic data was selected from 82 patients for radiomic models, including support vector machine (SVM) and logistic regression (LR) models. Furthermore, the nomogram incorporating radiomic signatures was constructed to evaluate its clinical efficacy. BTK was identified as an independent risk factor for HGGs through univariate and multivariate Cox regression analyses. Three radiomic features were selected to construct the SVM and LR models, and the validation set showed area under curve (AUCs) values of 0.711 (95% CI, 0.598–0.824) and 0.736 (95% CI, 0.627–0.844), respectively. The median survival times of the high Rad_score and low-Rad_score groups based on LR model were 15.53 and 23.03 months, respectively. In addition, the total risk score of each patient was used to construct a predictive nomogram, and the AUCs calculated from the corresponding time-dependent ROC curves were 0.533, 0.659, and 0.767 for 1, 3, and 5 years, respectively. BTK is an independent risk factor associated with poor prognosis in patients, and the radiomic model constructed in this study can effectively and non-invasively predict preoperative BTK expression levels and patient prognosis based on T1WI + C.








Similar content being viewed by others
Availability of Data and Materials
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding author/s.
Abbreviations
- HGG:
-
High-Grade Glioma
- BTK:
-
Bruton’s Tyrosine Kinase
- MRI:
-
Magnetic Resonance Imaging
- TCGA:
-
The Cancer Genome Atlas
- TCIA:
-
The Cancer Imaging Archive
- T1WI + C:
-
Contrast-Enhanced T1-Weighted Imaging
- CGGA:
-
Chinese Glioma Genome Atlas
- OS:
-
Overall survival
- GLRLM:
-
Gray-Level Run-Length Matrix
- GLSZM:
-
Gray-Level Size Zone Matrix
- GLDM :
-
Gray-Level Dependence Matrix
- GLCM:
-
Gray-Level Co-Occurrence Matrix
- NGTDM:
-
Neighboring Gray Tone Difference Matrix
- mRMR:
-
Maximum Relevance Minimum Redundancy
- RFE:
-
Recursive Feature Elimination
- SVM:
-
Support Vector Machine
- LR:
-
Logistic Regression
- ROC:
-
Receiver Operating Characteristic
- AUC:
-
Area Under the Curve
- DCA:
-
Decision Curve Analysis
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
References
Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15 Suppl 2(Suppl 2):ii1–56. https://doi.org/10.1093/neuonc/not151
Hu LS, Hawkins-Daarud A, Wang L, Li J, Swanson KR. Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett. 2020;477:97-106.https://doi.org/10.1016/j.canlet.2020.02.025
Hernández Martínez A, Madurga R, García-Romero N, Ayuso-Sacido Á. Unravelling glioblastoma heterogeneity by means of single-cell RNA sequencing. Cancer Lett. 2022;527:66-79. https://doi.org/10.1016/j.canlet.2021.12.008
Weller M, Wick W, Aldape K, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017.https://doi.org/10.1038/nrdp.2015.17
Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803-820. https://doi.org/10.1007/s00401-016-1545-1
Bush NAO, Butowski N. The Effect of Molecular Diagnostics on the Treatment of Glioma. Curr Oncol Rep. 2017;19(4):26. https://doi.org/10.1007/s11912-017-0585-6
Ma R, Taphoorn MJB, Plaha P. Advances in the management of glioblastoma. J Neurol Neurosurg Psychiatry. 2021;92(10):1103-1111. https://doi.org/10.1136/jnnp-2020-325334
Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434-452. https://doi.org/10.1038/nrclinonc.2016.204
Qin T, Mullan B, Ravindran R, et al. ATRX loss in glioma results in dysregulation of cell-cycle phase transition and ATM inhibitor radio-sensitization. Cell Rep. 2022;38(2):110216. https://doi.org/10.1016/j.celrep.2021.110216
Nguyen HN, Lie A, Li T, et al. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 2017;19(3):394-404. https://doi.org/10.1093/neuonc/now189
Huang LE. Impact of CDKN2A/B Homozygous Deletion on the Prognosis and Biology of IDH-Mutant Glioma. Biomedicines. 2022;10(2):246. https://doi.org/10.3390/biomedicines10020246
Reinhardt A, Stichel D, Schrimpf D, et al. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol. 2018;136(2):273-291. https://doi.org/10.1007/s00401-018-1837-8
E CC, J C. Bruton’s tyrosine kinase inhibitors: a promising emerging treatment option for multiple sclerosis. Expert opinion on emerging drugs. 2020;25(4). https://doi.org/10.1080/14728214.2020.1822817
Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32-42. https://doi.org/10.1056/NEJMoa1215637
Burger JA, Wiestner A. Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nat Rev Cancer. 2018;18(3):148-167. https://doi.org/10.1038/nrc.2017.121
Byrd JC, Furman RR, Coutre SE, et al. Ibrutinib Treatment for First-Line and Relapsed/Refractory Chronic Lymphocytic Leukemia: Final Analysis of the Pivotal Phase Ib/II PCYC-1102 Study. Clin Cancer Res. 2020;26(15):3918-3927. https://doi.org/10.1158/1078-0432.CCR-19-2856
Butler M, van Ingen Schenau DS, Yu J, et al. BTK inhibition sensitizes acute lymphoblastic leukemia to asparaginase by suppressing the amino acid response pathway. Blood. 2021;138(23):2383-2395. https://doi.org/10.1182/blood.2021011787
Tam CS, Anderson MA, Pott C, et al. Ibrutinib plus Venetoclax for the Treatment of Mantle-Cell Lymphoma. N Engl J Med. 2018;378(13):1211-1223. https://doi.org/10.1056/NEJMoa1715519
Lavitrano M, Ianzano L, Bonomo S, et al. BTK inhibitors synergise with 5-FU to treat drug-resistant TP53-null colon cancers. J Pathol. 2020;250(2):134-147. https://doi.org/10.1002/path.5347
Wang JD, Chen XY, Ji KW, Tao F. Targeting Btk with ibrutinib inhibit gastric carcinoma cells growth. Am J Transl Res. 2016;8(7):3003-3012.
Kim JM, Park J, Noh EM, et al. Bruton’s agammaglobulinemia tyrosine kinase (Btk) regulates TPA‑induced breast cancer cell invasion via PLCγ2/PKCβ/NF‑κB/AP‑1‑dependent matrix metalloproteinase‑9 activation. Oncol Rep. 2021;45(5):56. https://doi.org/10.3892/or.2021.8007
Yue C, Niu M, Shan QQ, et al. High expression of Bruton’s tyrosine kinase (BTK) is required for EGFR-induced NF-κB activation and predicts poor prognosis in human glioma. J Exp Clin Cancer Res. 2017;36(1):132. https://doi.org/10.1186/s13046-017-0600-7
Yang H, Liu X, Zhu X, et al. CPVL promotes glioma progression via STAT1 pathway inhibition through interactions with the BTK/p300 axis. JCI Insight. 2021;6(24):e146362. https://doi.org/10.1172/jci.insight.146362
Su YK, Bamodu OA, Su IC, et al. Combined Treatment with Acalabrutinib and Rapamycin Inhibits Glioma Stem Cells and Promotes Vascular Normalization by Downregulating BTK/mTOR/VEGF Signaling. Pharmaceuticals (Basel). 2021;14(9):876. https://doi.org/10.3390/ph14090876
Rt L, G D, D DR, P L, W van E. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. The British journal of radiology. 2017;90(1070). https://doi.org/10.1259/bjr.20160665
Bai HX, Lee AM, Yang L, et al. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89(1061):20151030. https://doi.org/10.1259/bjr.20151030
Li Y, Ammari S, Lawrance L, et al. Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion. Cancers (Basel). 2022;14(7):1778. https://doi.org/10.3390/cancers14071778
Niu L, Feng WH, Duan CF, Liu YC, Liu JH, Liu XJ. The Value of Enhanced MR Radiomics in Estimating the IDH1 Genotype in High-Grade Gliomas. Biomed Res Int. 2020;2020:4630218. https://doi.org/10.1155/2020/4630218
Y T, W M, Xc W, Gq Y, Rj G, H Z. Improving survival prediction of high-grade glioma via machine learning techniques based on MRI radiomic, genetic and clinical risk factors. European journal of radiology. 2019;120. https://doi.org/10.1016/j.ejrad.2019.07.010
Han W, Qin L, Bay C, et al. Deep Transfer Learning and Radiomics Feature Prediction of Survival of Patients with High-Grade Gliomas. AJNR Am J Neuroradiol. 2020;41(1):40-48. https://doi.org/10.3174/ajnr.A6365
Rui W, Ren Y, Wang Y, Gao X, Xu X, Yao Z. MR textural analysis on T2 FLAIR images for the prediction of true oligodendroglioma by the 2016 WHO genetic classification. J Magn Reson Imaging. 2018;48(1):74-83. https://doi.org/10.1002/jmri.25896
Liu X, Li Y, Qian Z, et al. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. NeuroImage: Clinical. 2018;20:1070–1077. https://doi.org/10.1016/j.nicl.2018.10.014
Zhao Z, Zhang KN, Wang Q, et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics, Proteomics & Bioinformatics. 2021;19(1):1-12. https://doi.org/10.1016/j.gpb.2020.10.005
Zhang K, Liu X, Li G, et al. Clinical management and survival outcomes of patients with different molecular subtypes of diffuse gliomas in China (2011–2017): a multicenter retrospective study from CGGA. Cancer Biology & Medicine. 2022;19(10):1460-1476. https://doi.org/10.20892/j.issn.2095-3941.2022.0469
Wang Y, Qian T, You G, et al. Localizing seizure-susceptible brain regions associated with low-grade gliomas using voxel-based lesion-symptom mapping. Neuro-Oncology. 2015;17(2):282-288. https://doi.org/10.1093/neuonc/nou130
Mayerhoefer ME, Materka A, Langs G, et al. Introduction to Radiomics. J Nucl Med. 2020;61(4):488-495. https://doi.org/10.2967/jnumed.118.222893
Lundberg SM, Erion G, Chen H, et al. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat Mach Intell. 2020;2(1):56-67. https://doi.org/10.1038/s42256-019-0138-9
He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. In: ; 2016:770–778. Accessed January 16, 2024. https://openaccess.thecvf.com/content_cvpr_2016/html/He_Deep_Residual_Learning_CVPR_2016_paper.html
Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015;115(3):211-252. https://doi.org/10.1007/s11263-015-0816-y
Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. Published online April 10, 2015. https://doi.org/10.48550/arXiv.1409.1556
Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773-782. https://doi.org/10.1038/s41587-019-0114-2
Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432-446. https://doi.org/10.1016/S0140-6736(18)30990-5
Jp F, Dc A. Current FDA-Approved Therapies for High-Grade Malignant Gliomas. Biomedicines. 2021;9(3). https://doi.org/10.3390/biomedicines9030324
Wang Q, Xiao F, Qi F, Song X, Yu Y. Risk Factors for Cognitive Impairment in High-Grade Glioma Patients Treated with Postoperative Radiochemotherapy. Cancer Res Treat. 2020;52(2):586-593. https://doi.org/10.4143/crt.2019.242
Kudus K, Wagner MW, Namdar K, et al. Increased confidence of radiomics facilitating pretherapeutic differentiation of BRAF-altered pediatric low-grade glioma. Eur Radiol. Published online October 7, 2023. https://doi.org/10.1007/s00330-023-10267-1
Lu J, Xu W, Chen X, Wang T, Li H. Noninvasive prediction of IDH mutation status in gliomas using preoperative multiparametric MRI radiomics nomogram: A mutlicenter study. Magn Reson Imaging. 2023;104:72-79. https://doi.org/10.1016/j.mri.2023.09.001
Wu Z, Yang Y, Zha Y. Radiomics Features on Magnetic Resonance Images Can Predict C5aR1 Expression Levels and Prognosis in High-Grade Glioma. Cancers (Basel). 2023;15(18):4661. https://doi.org/10.3390/cancers15184661
Schwartz GK, LoRusso PM, Dickson MA, et al. Phase I study of PD 0332991, a cyclin-dependent kinase inhibitor, administered in 3-week cycles (Schedule 2/1). Br J Cancer. 2011;104(12):1862-1868. https://doi.org/10.1038/bjc.2011.177
Brown JR, Eichhorst B, Hillmen P, et al. Zanubrutinib or Ibrutinib in Relapsed or Refractory Chronic Lymphocytic Leukemia. N Engl J Med. 2023;388(4):319-332. https://doi.org/10.1056/NEJMoa2211582
Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180-1191. https://doi.org/10.1016/S1470-2045(18)30413-3
Guo H, Xie M, Liu W, et al. Inhibition of BTK improved APAP-induced liver injury via suppressing proinflammatory macrophages activation by restoring mitochondrion function. Int Immunopharmacol. 2022;110:109036. https://doi.org/10.1016/j.intimp.2022.109036
Buggy JJ, Elias L. Bruton tyrosine kinase (BTK) and its role in B-cell malignancy. Int Rev Immunol. 2012;31(2):119-132. https://doi.org/10.3109/08830185.2012.664797
Shi W, Qu C, Wang X, Liang X, Tan Y, Zhang H. Diffusion kurtosis imaging combined with dynamic susceptibility contrast-enhanced MRI in differentiating high-grade glioma recurrence from pseudoprogression. Eur J Radiol. 2021;144:109941. https://doi.org/10.1016/j.ejrad.2021.109941
Shi Z, Jiang J, Xie L, Zhao X. Efficacy evaluation of contrast-enhanced magnetic resonance imaging in differentiating glioma from metastatic tumor of the brain and exploration of its association with patients’ neurological function. Front Behav Neurosci. 2022;16:957795. https://doi.org/10.3389/fnbeh.2022.957795
Chen S, Xu Y, Ye M, et al. Predicting MGMT Promoter Methylation in Diffuse Gliomas Using Deep Learning with Radiomics. J Clin Med. 2022;11(12):3445. https://doi.org/10.3390/jcm11123445
Joo L, Park JE, Park SY, et al. Extensive peritumoral edema and brain-to-tumor interface MRI features enable prediction of brain invasion in meningioma: development and validation. Neuro Oncol. 2021;23(2):324-333. https://doi.org/10.1093/neuonc/noaa190
Vickers AJ, Woo S. Decision curve analysis in the evaluation of radiology research. Eur Radiol. 2022;32(9):5787-5789. https://doi.org/10.1007/s00330-022-08685-8
Sugiura D, Maruhashi T, Okazaki IM, et al. Restriction of PD-1 function by cis-PD-L1/CD80 interactions is required for optimal T cell responses. Science. 2019;364(6440):558-566. https://doi.org/10.1126/science.aav7062
Li Y, Jia L, Kim JK, et al. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell. 2021;28(9):1597-1613.e7. https://doi.org/10.1016/j.stem.2021.04.011
Ning Z, Liu K, Xiong H. Roles of BTLA in Immunity and Immune Disorders. Front Immunol. 2021;12:654960. https://doi.org/10.3389/fimmu.2021.654960
Funding
This paper was funded by the National Key R&D Program of China, No. 2022YFF0608404.
Author information
Authors and Affiliations
Contributions
JW, ZDN, and MSC formulated the overall research design and conceived the concept. JCG made significant contributions to data collection and part of data processing. WX and SC completed the majority of data processing. JCG and ZDN drafted the manuscript. All authors made critical revision of the manuscript. All authors contributed equally to this paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
No animal, human, and potentially identifiable human images or data studies are presented in this manuscript.
Consent for Publication
All authors agreed.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, C., Sun, C., Wang, X. et al. BTK Expression Level Prediction and the High-Grade Glioma Prognosis Using Radiomic Machine Learning Models. J Digit Imaging. Inform. med. (2024). https://doi.org/10.1007/s10278-024-01026-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10278-024-01026-9