Abstract
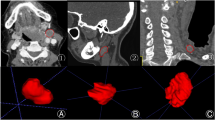
The purpose is to train and evaluate a deep learning (DL) model for the accurate detection and segmentation of abnormal cervical lymph nodes (LN) on head and neck contrast-enhanced CT scans in patients diagnosed with lymphoma and evaluate the clinical utility of the DL model in response assessment. This retrospective study included patients who underwent CT for abnormal cervical LN and lymphoma assessment between January 2021 and July 2022. Patients were grouped into the development (n = 76), internal test 1 (n = 27), internal test 2 (n = 87), and external test (n = 26) cohorts. A 3D SegResNet model was used to train the CT images. The volume change rates of cervical LN across longitudinal CT scans were compared among patients with different treatment outcomes (stable, response, and progression). Dice similarity coefficient (DSC) and the Bland–Altman plot were used to assess the model’s segmentation performance and reliability, respectively. No significant differences in baseline clinical characteristics were found across cohorts (age, P = 0.55; sex, P = 0.13; diagnoses, P = 0.06). The mean DSC was 0.39 ± 0.2 with a precision and recall of 60.9% and 57.0%, respectively. Most LN volumes were within the limits of agreement on the Bland–Altman plot. The volume change rates among the three groups differed significantly (progression (n = 74), 342.2%; response (n = 8), − 79.2%; stable (n = 5), − 8.1%; all P < 0.01). Our proposed DL segmentation model showed modest performance in quantifying the cervical LN burden on CT in patients with lymphoma. Longitudinal changes in cervical LN volume, as predicted by the DL model, were useful for treatment response assessment.






Similar content being viewed by others
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- DL:
-
Deep learning
- DSC:
-
Dice similarity coefficient
- HL:
-
Hodgkin lymphoma
- ICC:
-
Intraclass correlation coefficient
- LN:
-
Lymph node
- NHL:
-
Non-Hodgkin lymphoma
- RECIST:
-
Response Evaluation Criteria in Solid Tumors
References
Eisenhauer EA, Therasse P, Bogaerts J et al (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45:228-247
Cheson BD, Horning SJ, Coiffier B et al (1999) Report of an international workshop to standardize response criteria for non-Hodgkin's lymphomas. J Clin Oncol 17:1244-1244
Cheson BD (2008) Staging and evaluation of the patient with lymphoma. Hematol Oncol Clin North Am 22:825–837, vii-viii
Cheson BD, Fisher RI, Barrington SF et al (2014) Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol 32:3059
Wulff A, Fabel M, Freitag-Wolf S et al (2013) Volumetric response classification in metastatic solid tumors on MSCT: initial results in a whole-body setting. Eur J Radiocol 82:e567-e573
Takahashi Y, Takashima S, Hakucho T et al (2013) Diagnosis of regional node metastases in lung cancer with computer-aided 3D measurement of the volume and CT-attenuation values of lymph nodes. Acad Radiol 20:740-745
Basilio-de-Leo CI, Villeda-Sandoval CI, Culebro-García C, Rodríguez-Covarrubias F, Castillejos-Molina RA (2015) Volumetric assessment of lymph node metastases in patients with non-seminomatous germ cell tumours treated with chemotherapy. Can Urol Assoc J 9:E247
Fabel M, von Tengg-Kobligk H, Giesel F et al (2008) Semi-automated volumetric analysis of lymph node metastases in patients with malignant melanoma stage III/IV-A feasibility study. Eur Radiol 18:1114-1122
Kim T, Lee KH, Ham S et al (2020) Active learning for accuracy enhancement of semantic segmentation with CNN-corrected label curations: Evaluation on kidney segmentation in abdominal CT. Sci Rep 10:1-7
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Editor, (ed)^(eds) Book 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. (Series 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation). Springer International Publishing, Cham, pp 424–432
Courot A, Cabrera DL, Gogin N et al (2021) Automatic cervical lymphadenopathy segmentation from CT data using deep learning. Diagn Interv Imaging 102:675-681
Tekchandani H, Verma S, Londhe ND, Jain RR, Tiwari A (2022) Computer aided diagnosis system for cervical lymph nodes in CT images using deep learning. Biomed Signal Process Control 71:103158
Cardenas CE, Beadle BM, Garden AS et al (2021) Generating high-quality lymph node clinical target volumes for head and neck cancer radiation therapy using a fully automated deep learning-based approach. Int J Radiat Oncol Biol Phys 109:801–812
Taku N, Wahid KA, van Dijk LV et al (2022) Auto-detection and segmentation of involved lymph nodes in HPV-associated oropharyngeal cancer using a convolutional deep learning neural network. Clin Transl Radiat Oncol 36:47-55
Myronenko A (2019) 3D MRI brain tumor segmentation using autoencoder regularization. In: Editor, (ed)^(eds) Book 3D MRI brain tumor segmentation using autoencoder regularization. (Series 3D MRI brain tumor segmentation using autoencoder regularization). Springer, pp 311–320
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297-302
Shafiei A, Bagheri M, Farhadi F et al (2021) CT evaluation of lymph nodes that merge or split during the course of a clinical trial: limitations of RECIST 1.1. Radiol Imaging Cancer 3:e200090
Weisman AJ, Kieler MW, Perlman SB et al (2020) Convolutional neural networks for automated PET/CT detection of diseased lymph node burden in patients with lymphoma. Radiol Artificial Intelligence 2:e200016
Shin H-C, Roth HR, Gao M et al (2016) Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35:1285-1298
Funding
This work was supported by a Research Fund from Taejoon Pharmaceutical Co., Ltd., Korea, and by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2021R1I1A1A01040285, 2020R1F1A1070517). The funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
Yoonho Nam wrote the methods section of the paper and performed the deep learning analysis; Su-Youn Kim contributed to deep learning analysis; Kyu-Ah Kim contributed to deep learning analysis; Euna Kwon contributed to deep learning analysis; Yoo Hyun Lee collected the data; Jinhee Jang supervised the project; Min Kyoung Lee contributed data; Yangsean Choi conceived and designed the analysis, performed the statistical analysis, wrote the paper, and acquired the fund.
Corresponding author
Ethics declarations
Ethics Approval
The institutional review boards of Seoul St. Mary’s Hospital and Yeouido St. Mary’s Hospital approved this retrospective study and waived the need for informed consent.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Points
• DL model showed modest segmentation performance of cervical LN in lymphoma patients.
• The ground truth and predicted LN volumes were within the limits of the agreement.
• Predicted longitudinal cervical LN volume changes were useful for treatment response assessment.
Critical Relevance Statement
Deep learning-based segmentation of cervical lymphoma burden on CT would be useful for radiologic treatment response assessment.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nam, Y., Kim, SY., Kim, KA. et al. Development and Validation of Deep Learning-Based Automated Detection of Cervical Lymphadenopathy in Patients with Lymphoma for Treatment Response Assessment: A Bi-institutional Feasibility Study. J Digit Imaging. Inform. med. 37, 734–743 (2024). https://doi.org/10.1007/s10278-024-00966-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-024-00966-6