Abstract
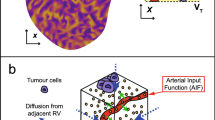
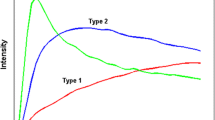
Pharmacokinetic (PK) parameters, revealing changes in the tumor microenvironment, are related to the pathological information of breast cancer. Tracer kinetic models (e.g., Tofts-Kety model) with a nonlinear least square solver are commonly used to estimate PK parameters. However, the method is sensitive to noise in images. To relieve the effects of noise, a deconvolution (DEC) method, which was validated on synthetic concentration–time series, was proposed to accurately calculate PK parameters from breast dynamic contrast-enhanced magnetic resonance imaging. A time-to-peak-based tumor partitioning method was used to divide the whole tumor into three tumor subregions with different kinetic patterns. Radiomic features were calculated from the tumor subregion and whole tumor-based PK parameter maps. The optimal features determined by the fivefold cross-validation method were used to build random forest classifiers to predict molecular subtypes, Ki-67, and tumor grade. The diagnostic performance evaluated by the area under the receiver operating characteristic curve (AUC) was compared between the subregion and whole tumor-based PK parameters. The results showed that the DEC method obtained more accurate PK parameters than the Tofts method. Moreover, the results showed that the subregion-based Ktrans (best AUCs = 0.8319, 0.7032, 0.7132, 0.7490, 0.8074, and 0.6950) achieved a better diagnostic performance than the whole tumor-based Ktrans (AUCs = 0.8222, 0.6970, 0.6511, 0.7109, 0.7620, and 0.5894) for molecular subtypes, Ki-67, and tumor grade. These findings indicate that DEC-based Ktrans in the subregion has the potential to accurately predict molecular subtypes, Ki-67, and tumor grade.






Similar content being viewed by others
Data Availability
The data that support the findings of this study are available upon request from the authors.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A: Cancer statistics, 2022. CA Cancer J Clin 72:7-33, 2022
Huber KE, Carey LA, Wazer DE: Breast cancer molecular subtypes in patients with locally advanced disease: impact on prognosis, patterns of recurrence, and response to therapy. Semin Radiat Oncol 19:204-210, 2009
Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA: Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol 11:174-183, 2010
Rakha EA, et al.: Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res 12:207, 2010
Fan M, et al.: A framework for deep multitask learning with multiparametric magnetic resonance imaging for the joint prediction of histological characteristics in breast cancer. IEEE Journal of Biomedical Health Informatics 26:3884-3895, 2022
Reig B, Heacock L, Geras KJ, Moy L: Machine learning in breast MRI. J Magn Reson Imaging 52:998-1018, 2020
Thomas AA, et al.: Dynamic contrast enhanced T1 MRI perfusion differentiates pseudoprogression from recurrent glioblastoma. J Neurooncol 125:183-190, 2015
Yankeelov TE, Gore JC: Dynamic contrast enhanced magnetic resonance imaging in oncology: theory, data acquisition, analysis, and examples Curr Med Imaging Rev 3:91–107, 2007
Elshafeey N, et al.: Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma. Nat Commun 10:3170, 2019
Ioannidis GS, Maris TG, Nikiforaki K, Karantanas A, Marias K: Investigating the correlation of Ktrans with semi-quantitative MRI parameters towards more robust and reproducible perfusion imaging biomarkers in three cancer types. IEEE J Biomed Health Inform 23:1855-1862, 2019
Kang SR, Kim HW, Kim HS: Evaluating the relationship between dynamic contrast-enhanced MRI (DCE-MRI) parameters and pathological characteristics in breast cancer. J Magn Reson Imaging 52:1360-1373, 2020
Tofts PS, et al.: Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 10:223-232, 1999
Brix G, Semmler W, Port R, Schad LR, Layer G, Lorenz WJ: Pharmacokinetic parameters in CNS Gd-DTPA enhanced MR imaging. J Comput Assist Tomogr 15:621, 1991
Tofts PS, Kermode AG: Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn Reson Med 17:357–367, 1991
Fang K, et al.: Convolutional neural network for accelerating the computation of the extended Tofts model in dynamic contrast-enhanced magnetic resonance imaging. J Magn Reson Imaging 53:1898-1910, 2021
Ulas C, et al.: Convolutional neural networks for direct inference of pharmacokinetic parameters: application to stroke dynamic contrast-enhanced MRI. Front Neurol 9:1147, 2018
Makkat S, Luypaert R, Sourbron S, Stadnik T, De Mey J: Quantification of perfusion and permeability in breast tumors with a deconvolution‐based analysis of second‐bolus T1‐DCE data. J Magn Reson Imaging 25:1159-1167, 2007
Makkat S, et al.: Deconvolution-based dynamic contrast-enhanced MR imaging of breast tumors: correlation of tumor blood flow with human epidermal growth factor receptor 2 status and clinicopathologic findings—preliminary results. Radiol Manage 249:471-482, 2008
Li ZW, et al.: Application of whole-lesion histogram analysis of pharmacokinetic parameters in dynamic contrast-enhanced MRI of breast lesions with the CAIPIRINHA-Dixon-TWIST-VIBE technique. J Magn Reson Imaging 47:91-96, 2018
Banaie M, Soltanian-Zadeh H, Saligheh-Rad HR, Gity M: Spatiotemporal features of DCE-MRI for breast cancer diagnosis. Comput Methods Programs Biomed 155:153-164, 2018
Thibault G, et al.: DCE-MRI texture features for early prediction of breast cancer therapy response. Tomography 3:23-32, 2017
Machireddy A, et al.: Early prediction of breast cancer therapy response using multiresolution fractal analysis of DCE-MRI parametric maps. Tomography 5:90-98, 2019
Nagasaka K, Satake H, Ishigaki S, Kawai H, Naganawa S: Histogram analysis of quantitative pharmacokinetic parameters on DCE-MRI: correlations with prognostic factors and molecular subtypes in breast cancer. Breast Cancer 26:113-124, 2019
Monti S, et al.: DCE-MRI pharmacokinetic-based phenotyping of invasive ductal carcinoma: a radiomic study for prediction of histological outcomes. Contrast Media Mol Imaging 2018:5076269, 2018
Fan M, et al.: Radiomic analysis of imaging heterogeneity in tumours and the surrounding parenchyma based on unsupervised decomposition of DCE-MRI for predicting molecular subtypes of breast cancer. Eur Radiol 29:4456-4467, 2019
Fan M, et al.: DCE-MRI texture analysis with tumor subregion partitioning for predicting Ki-67 status of estrogen receptor-positive breast cancers. J Magn Reson Imaging 48:237-247, 2018
Fan M, et al.: Tumour heterogeneity revealed by unsupervised decomposition of dynamic contrast-enhanced magnetic resonance imaging is associated with underlying gene expression patterns and poor survival in breast cancer patients. Breast Cancer Res 21:112, 2019
Fan M, Xia PP, Clarke R, Wang Y, Li LH: Radiogenomic signatures reveal multiscale intratumour heterogeneity associated with biological functions and survival in breast cancer. Nat Commun 11:4861, 2020
Wu J, Gong GH, Cui Y, Li RJ: Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reson Imaging 44:1107-1115, 2016
Chitalia RD, et al.: Imaging phenotypes of breast cancer heterogeneity in preoperative breast dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) scans predict 10-year recurrence. Clin Cancer Res 26:862-869, 2020
Wu J, et al.: Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology 288:26-35, 2018
Carey LA, et al.: Race, breast cancer subtypes, and survival in the carolina breast cancer study. Jama the Journal of the American Medical Association 295:2492-2502, 2006
Fan M, et al.: Joint prediction of breast cancer histological grade and Ki-67 expression level based on DCE-MRI and DWI radiomics. IEEE J Biomed Health Inform 24:1632-1642, 2020
Liao GJ, et al.: Background parenchymal enhancement on breast MRI: A comprehensive review. J Magn Reson Imaging 51:43-61, 2020
Chuang KS, Tzeng HL, Chen S, Wu J, Chen TJ: Fuzzy c-means clustering with spatial information for image segmentation. Comput Med Imaging Graph 30:9-15, 2006
Wu O, Ostergaard L, Weisskoff RM, Benner T, Rosen BR, Sorensen AG: Tracer arrival timing-insensitive technique for estimating flow in MR perfusion-weighted imaging using singular value decomposition with a block-circulant deconvolution matrix. Magn Reson Med 50:164-174, 2003
Ho KC, Scalzo F, Sarma KV, El-Saden S, Arnold CW: A temporal deep learning approach for MR perfusion parameter estimation in stroke. 2016 23rd International Conference on Pattern Recognition (ICPR) 2016:1315–1320, 2016
Winther HB, et al.: Deep semantic lung segmentation for tracking potential pulmonary perfusion biomarkers in chronic obstructive pulmonary disease (COPD): the multi‐ethnic study of atherosclerosis COPD study. J Magn Reson Imaging 51:571-579, 2020
DiCarlo JC, et al.: Analysis of simplicial complexes to determine when to sample for quantitative DCE MRI of the breast. Magn Reson Med 89:1134-1150, 2023
de Bazelaire C, et al.: Accuracy of perfusion MRI with high spatial but low temporal resolution to assess invasive breast cancer response to neoadjuvant chemotherapy: a retrospective study. BMC Cancer 11:361, 2011
Rosa E, Sima DM, Menze B, Kirschke JS, Robben D: AIFNet: automatic vascular function estimation for perfusion analysis using deep learning. Med Image Anal 74:102211, 2021
Shao J, et al.: DCE-MRI pharmacokinetic parameter maps for cervical carcinoma prediction. Comput Biol Med 118:103634, 2020
Wake N, et al.: Accuracy and precision of quantitative DCE-MRI parameters: how should one estimate contrast concentration? Magn Reson Imaging 52:16-23, 2018
Torheim T, et al.: Cluster analysis of dynamic contrast enhanced MRI reveals tumor subregions related to locoregional relapse for cervical cancer patients. Acta Oncol 55:1294-1298, 2016
Lu HC, Yin JD: Texture analysis of breast DCE-MRI based on intratumoral subregions for predicting HER2 2+ status. Front Oncol 10:543, 2020
van Griethuysen JJM, et al.: Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104-e107, 2017
Zhang L, Fan M, Wang S, Xu M, Li L: Radiomic analysis of pharmacokinetic heterogeneity within tumor based on the unsupervised decomposition of dynamic contrast-enhanced MRI for predicting histological characteristics of breast cancer. J Magn Reson Imaging 55:1636-1647, 2022
Barnes SR, Ng TS, Santa-Maria N, Montagne A, Zlokovic BV, Jacobs RE: ROCKETSHIP: a flexible and modular software tool for the planning, processing and analysis of dynamic MRI studies. BMC Med Imaging 15:19, 2015
Whitcher B, Schmid VJ: Quantitative analysis of dynamic contrast-enhanced and diffusion-weighted magnetic resonance imaging for oncology in R. Journal of Statistical Software 44:1-29, 2011
Xie TW, et al.: Machine learning-based analysis of MR multiparametric radiomics for the subtype classification of breast cancer. Front Oncol 9:505, 2019
Heisen M, Fan X, Buurman J, van Riel NA, Karczmar GS, ter Haar Romeny BM: The influence of temporal resolution in determining pharmacokinetic parameters from DCE‐MRI data. Magn Reson Med 63:811-816, 2010
Matsukuma M, et al.: The kinetic analysis of breast cancer: An investigation of the optimal temporal resolution for dynamic contrast-enhanced MR imaging. Clin Imaging 61:4-10, 2020
Funding
This study was supported in part by grants from the National Key R&D Program of China (2018YFA0701700, 2021YFE0203700), the National Natural Science Foundation of China under Grant (62271178, U21A20521, U1809205, and 62302014), and the Natural Science Foundation of Zhejiang Province of China under Grant (LR23F010002).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics Approval
This retrospective study was approved by the Institutional Review Board of the First Affiliated Hospital of Zhejiang Chinese Medical University.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Competing Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Fan, M. & Li, L. Deconvolution-Based Pharmacokinetic Analysis to Improve the Prediction of Pathological Information of Breast Cancer. J Digit Imaging. Inform. med. 37, 13–24 (2024). https://doi.org/10.1007/s10278-023-00915-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-023-00915-9