Abstract
The main purpose of this study is to analyze the intrinsic tumor physiologic characteristics in patients with sarcoma through model-free analysis of dynamic contrast enhanced MR imaging data (DCE-MRI). Clinical data were collected from three patients with two different types of histologically proven sarcomas who underwent conventional and advanced MRI examination prior to excision. An advanced matrix factorization algorithm has been applied to the data, resulting in the identification of the principal time-signal uptake curves of DCE-MRI data, which were used to characterize the physiology of the tumor area, described by three different perfusion patterns i.e. hypoxic, well-perfused and necrotic one. The performance of the algorithm was tested by applying different initialization approaches with subsequent comparison of their results. The algorithm was proven to be robust and led to the consistent segmentation of the tumor area in three regions of different perfusion, i.e. well-perfused, hypoxic and necrotic. Results from the model-free approach were compared with a widely used pharmacokinetic (PK) model revealing significant correlations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Hypoxia is a hallmark of most solid malignant neoplasms and is usually related with more aggressive tumor phenotypes and resistance to chemotherapy and radiotherapy [7, 15]. Tumors are associated with abnormal rapid formation of new blood vessels, in an aim to overcome the increased oxygen and other nutrient demands during tumor growth. However, tumor newly-formed vessels are typically dysfunctional impeding proper vessel-to-tissue delivery of both oxygen and chemotherapy drugs. Eventually, the augmented interstitial fluid pressure is followed by a subsequent decrease of tumor perfusion, further favoring tumor hypoxia [5]. As a result, hypoxia estimation from imaging data can play a key role in tumor characterization and lead to a more accurate and effective treatment planning.
A number of different techniques have been used for the assessment of tumor hypoxia [29]. These include invasive techniques, such as pO2 electrode measurements and immunohistochemistry of exogenous markers e.g. pimonidazole (PIMO). Imaging techniques have also been widely used, including positron emission tomography (PET) imaging using radioactive hypoxia tracers and MR imaging. In particular, dynamic contrast enhanced MR Imaging (DCE-MRI) is a non-invasive perfusion imaging technique, which offers high spatial resolution, can be performed on clinical MRI scanners with standard specifications and can yield information concerning tissue oxygenation and vascularization. It has been used in previous studies for different purposes, such as for determining tumor grading, predicting response to therapy and differentiating between benign and malignant lesions [4, 21]. DCE-MRI involves the intravenous injection of a contrast agent (CA) with T1-weighted (w) sequences acquired before, during and after injection. The paramagnetic CA diffuses from the arteries to the extravascular extracellular space (EES), which accelerates the T1 relaxation process of tissue and leads to signal enhancement in T1-w MR images. As mentioned previously, hypoxic tumors are typically characterized by poor perfusion, thus DCE-MRI could provide useful information about the presence and distribution of hypoxia in tumors.
Currently, compartmental pharmacokinetic (PK) modeling is the most widely used technique for the DCE-MRI data analysis. It relies on dividing the anatomic area of interest in two compartments, the intravascular and EES. Perfusion related parameters are estimated in order to quantify the CA transfer rates between the different compartments and the percentage volume they occupy. However, PK models involve a number of limitations such as the signal to contrast conversion and the computationally expensive fitting algorithms [3].
A different model-free approach that has been used for the analysis of DCE-MRI data, comprises of the shape classification of the time-signal uptake curves of image voxels in a selected region of interest (ROI) within the tumor. According to the literature [12], the most common enhancement patterns of the DCE time-signal curves are three (Fig. 1). The first one is characterized by steady enhancement of the MR signal intensity (Type 1), the second one by enhancement followed by some wash-out (Type 2) and the third one by fast enhancement and rapid wash-out (Type 3). The analysis of the structure of the DCE curves is somewhat simplistic to be used for characterizing the tumor physiology. However, if combined with pattern recognition (PR) techniques, it is able to provide an automatic identification of the enhancement patterns that characterize the DCE-MRI data. Model-free and model-based methods applied on DCE-MRI data have been reviewed previously in [3].
In a previous study [2], DCE-MRI data of a rat prostate tumor were analyzed using PK modeling, resulting in the identification of well-perfused and necrotic areas, which was further validated by histology. However, PET imaging was also used for the identification of hypoxic areas. In [6, 23], DCE-MRI prostate cancer data have been analyzed using two different PR methods, nonnegative matrix factorization (NMF) and Gaussian Mixture Models (GMMs) respectively, leading to a three-area classification of the tumor in well-perfused, necrotic and hypoxic regions. The results were correlated with hypoxic areas defined by the hypoxia marker PIMO, necrotic areas defined by hematoxylin-eosin (H&E) and well-perfused areas derived from the Hoffmann pharmacokinetic model [8]. In [24], DCE pharmacokinetic parameters (kep, ktrans, ve) have been compared with histopathological parameters (microvessel density parameters and tumor proliferation index) in head and neck squamous cell carcinoma (HNSCC) resulting in important correlations. In [10], the DCE parameter kep was proven to be closely correlated with VEGF expression in breast tumors. DCE-MRI parameters have been also correlated with pimonidazole and CA9 staining in patients with head-and-neck cancer [17], indicating that they can be used as surrogate markers of hypoxia. In [31], semi-quantitative parameters of DCE-MRI were compared with H&E, PIMO and vascular endothelial growth factor (VEGF) immunohistochemistry in a maxillofacial VX2 rabbit model. Important correlations were revealed, showing that DCE-MRI is an attractive choice to offer a reliable estimation of tumor hypoxia.
Since such studies strongly indicate that DCE-MRI parameters convey hypoxia information, it is important to try to define automatic methods to extract hypoxia maps from DCE-MRI functional images. Design of a rapid and accurate image analysis algorithm for DCE-MRI data may have great impact on the evaluation of drug delivery as well as the development of specific treatments with antiangiogenic agents. To this end, herein we present a machine learning, model-free method that automatically decomposes the image in three dominant components corresponding to necrotic, hypoxic and well-perfused areas.
In particular, in this study we extended the work in [23] by applying an advanced version of the NMF algorithm and by testing it on human patient data. In particular, the block coordinate update NMF (BU-NMF) algorithm, a PR technique, was applied to DCE-MRI data derived from patients with histologically proven sarcomas. The algorithm detects the most representative patterns that comprise the data. In our previous studies [27, 28] we have examined the efficiency and robustness of this algorithm on a single patient and investigated the most suitable initialization approach to use. In the present study, the BU-NMF algorithm is tested on three patients, which have different types of sarcomas. Furthermore, in contrast to our previous reported works [27, 28], where a single slice was used, in this work information from the entire tumor volume was given as input so that the maximum available information regarding the malignancy could be used. The proposed BU-NMF-based method was applied using three different initialization approaches and the structures of the patterns identified seemed to approximate the three most common shapes of the DCE main signal-time curves (Types 1, 2 and 3), which were described previously. Well-perfused areas were defined as those having the Type 3 curve shape, hypoxic ones were assigned to the Type 2 shape and necrotic regions were those with enhancement profile similar to Type 1 curve. The classification of tumor areas has also been used in other studies investigating the role of DCE-MRI in defining hypoxia [9]. The proposed approach could be considered as a quantitative morphological analysis of the examined structures. In addition, we compare the spatial maps extracted by the PR technique with the parametric maps derived from the pharmacokinetic Extended Tofts Model (ETM), and investigate the correlation of the two different perfusion mappings for the selected tumor areas. In the ktrans map derived from the ETM model and for comparison purposes, image areas with high ktrans were labelled as well-perfused areas, those with ktrans zero or close to zero as necrotic areas, and those with intermediate values as hypoxic regions. Histopathology results provided by experts are also supporting, at a certain extent, the obtained results. This approach involves different types of analysis and has the advantage that by combining all the extracted information, a more global view of the tumor physiology characteristics can be obtained.
2 Materials and methods
2.1 Description of dataset
Three male patients, with median age 58 years, were included in the study. All of them were diagnosed with sarcomas, and more specifically one lower limb malignant peripheral nerve sheath tumor (MPNST), one thigh pleomorphic liposarcoma and one neck pleomorphic liposarcoma. This paper refers to the lower limb MPNST patient as Patient #1 while the patient with thigh pleomorphic liposarcoma is mentioned as Patient #2 and the patient with neck pleomorphic liposarcoma as Patient #3. All data were provided anonymously by the University Hospital of Heraklion (Pa.G.N.I.). The patients underwent conventional and advanced MRI prior to excision. The DCE-MRI experiments were performed on a 1.5 T Siemens scanner using the contrast agent gadopentetate dimeglumine (Gd-DTPA) at routinely used dose. For the acquisition of T1-w MRI sequences, six separate acquisitions were made prior to the CA injection, with variable flip angles (VFA) of 5°, 10°, 15°, 20°, 30° and 60°. T1-w MR images were acquired with fast three-dimensional spoiled gradient echo (SPGR) with the following parameters: TR = 7 ms, TE = 3.23 ms, flip angle = 15°, voxel size 1.04 × 1.65 × 5 mm for Patient #1 and Patient #3 and 1.15 × 1.82 × 5 mm for Patient #2, acquisition matrix 192 × 121, number of slices = 14, 45 time points for DCE-MRI protocol and temporal resolution 6.44 s. DCE datasets were co-registered to the arterial phase (maximum signal-to-noise ratio (SNR)).
2.2 Pharmacokinetic model approach
The PK model used in our analysis is the ETM, which is a modified version of the well-known Tofts model (TM) [26]. TM produces reliable results only when the tissue is weakly vascularized, whereas the ETM can also be applied to highly perfused tumors [22]. The contrast agent leaves the plasma space and enters the EES at a rate represented by ktrans and returns by kep = ktrans/ve, which is the exchange rate from EES to the plasma space where ve is the volume of EES. Both ktrans and kep are measured in [min-1]. In this study, the arterial input function (AIF) was calculated from a large vessel within the region of imaging and for the signal to contrast conversion, the method of variable flip angles (VFA) was applied. The parametric map of the ktrans rate was used not only for the initialization of the PR approach but also for comparing the results of the PR method with the results of the PK model method.
2.3 Pattern recognition approach
Images often contain redundant information since adjacent pixels in an image are highly correlated. To this end, dimensionality reduction algorithms are typically used to extract the important information from the image, while incurring very little error. Matrix factorization and principal component analysis are among the most popular methods for data representation in a lower-rank space. NMF, due to its non-negativity constraints, is distinguished from other matrix factorization methods such as principal component analysis (PCA). NMF results have more obvious visual interpretation and since it allows only additive combinations, it is suitable for uniting parts to generate a whole, leading to parts-based representations [13]. In our work, we want to identify certain patterns associated with cancer physiology and NMF learns to represent our MRI data as a linear combination of basis images, called patterns, each of them carrying a different weight. In this way, we can identify the pattern followed by each voxel as well as the contribution of the other patterns to each voxel of the image. This allows for a visual representation, which qualitatively resembles the nature of cancer behavior.
The basic NMF problem consists in finding an approximate decomposition of a large dimension data matrix A of size m x n into two low-rank nonnegative matrices, m x k matrix W and k x n matrix H. The ultimate aim is to minimize the functional:
The value of k is usually selected such that k < < min (m, n) which confirms the fact that the product WH, named the NMF of matrix A, equivalents to a compressed form of A. In practice, it corresponds to the number of basic patterns that NMF is going to use in order to represent the data in A.
The minimization of Eq. (1) involves some significant challenges. It implies the lack of convexity in both W and H, thus the existence of local minima. In addition, there is no unique solution of the minimization problem in Eq. (1) since the solution matrices W and H could also be replaced by an infinite number of other solution pairs, such as WS and S−1H for any nonnegative invertible matrix S having a nonnegative inverse S−1.
The NMF problem has been approached by several numerical methods resulting in different solutions [1]. Lee and Seung [14] established the first well-known NMF algorithm that is based on multiplicative update rules in order to minimize the Euclidean distance described in Eq. (1). Particularly, it can be shown that the square Euclidean distance measure used in Eq. (1) is nonincreasing under the iterative updated rules described in Algorithm 1, which is presented below [1].
Algorithm 1 – Multiplicative update
W = rand(m, k); % initialize W as random dense matrix
H = rand(k, n); % initialize H as random dense matrix
for i = 1: maxiter
H = H. * (WTA)./(WTWH + 10−9);
W = W. * (AHT)./(WHHT + 10−9);
end
Alternating least squares (ALS) algorithms [18] are significantly faster to converge with respect to the multiplicative update algorithms and are based on the fact that while the minimization problem in Eq. (1) is not convex in both W and H, it is convex in either W or H. The update rules used are presented in Algorithm 2 [1].
Algorithm 2 – Alternating least squares
W = rand(m, k); % initialize W as random dense matrix or use another initialization
for i = 1: maxiter
(LS) Solve for H in matrix equation WTWH = WTA.
(NONNEG) Set all negative elements in H to 0.
(LS) Solve for W in matrix equation HHTWT = HAT.
(NONNEG) Set all negative elements in W to 0.
end
In our work, we opted for the BU-NMF algorithm, which is an efficient algorithm with simple update steps and offering global convergence under certain assumptions [30]. It relies on regularized multi-convex optimization, which is a method used in problems characterized by non-convexity and non-smoothness.
A set of points is called block multiconvex if its projection to each block of variables is convex but can be generally nonconvex. We consider a variable x, which consists of s blocks (x1, … , x s ), a set X that is a closed and block multiconvex subset of ℝn, f is a differentiable and block multiconvex function, and (r1, … , r s ) are extended value convex functions. The optimization problem is described by the following equation [30]:
Below it is presented the BU-NMF algorithm we used for our analysis by choosing one of the three proposed choices of update schemes in [30].
Algorithm 3 – block-coordinate update method
Initialization: choose two initial points \( \left({x}_1^{-1},\dots, {x}_s^{-1}\right)=\left({x}_1^0,\dots, {x}_s^{0.}\right) \)
for k = 1 , 2 , … do
for i = 1 , 2 , … , s do
\( {x}_i^k\leftarrow \underset{\mathrm{x}\upepsilon \mathrm{X}}{\mathrm{argmin}}\left\langle {\widehat{g}}_i^k,{x}_i-{\widehat{x}}_i^{k-1}\right\rangle +\frac{L_i^{k-1}}{2}{\left\Vert {x}_i-{\widehat{x}}_i^{k-1}\right\Vert}^2+{r}_i\left({x}_i\right) \) (update scheme)
end for
if stopping criteria is satisfied then
return \( \left({x}_1^k,\dots, {x}_s^k\right) \)
end if
end for
In the update scheme formula used in Algorithm 3, \( {\widehat{x}}_i^{k-1} \) denotes an extrapolated point, \( {\widehat{g}}_i^k \) is the block-partial gradient of f at \( {\widehat{x}}_i^{k-1} \) and \( {L}_i^{k-1} \) is the Lipschitz constant of \( \nabla {f}_i^k \).
In [30] it was shown that the BU-NMF algorithm, when tested on synthetic, hyperspectral and real image datasets, it gave faster and more accurate results compared to other algorithms, such as multiplicative update, ALS, alternating direction method (ADM) and Blockpivot.
All the numerical results presented in this work have been obtained by Matlab 8.1.0.604 (R2013a) implemented on an Intel Core 2 i7–4770 processor, 3.4 GHz with 16 GB RAM. The code for the BU-NMF algorithm was obtained from [30]. The ETM pharmacokinetic results were obtained using a dedicated software tool for DCE-MRI data analysis [11].
2.3.1 Data pre-processing
The pre-contrast phase of the MR signal was first removed from the raw signal. A small number of pre-contrast acquisitions were obtained in order to observe the effect of the CA to MR signal intensity. Afterwards, the baseline was removed in order to extract the net effect of CA uptake on the signal intensity.
Finally, the smoothing spline method was implemented in order to remove the noise from the dynamic DCE-MRI data and obtain a smoother signal. In particular, a cubic smoothing spline algorithm was applied to the data for fitting a smooth curve to the noisy MR signal. In Fig. 2, we can see the plots resulting from all the previously described pre-processing steps applied to the original intensity curve of a specific image pixel from Patient #1.
2.3.2 BU-NMF initialization
Two different approaches have been tested for the initialization of the BU-NMF algorithm:
-
1.
Random initialization: The matrices W and H are typically initialized with random nonnegative values in the standard NMF algorithm as shown in the three algorithms that were formerly presented. In our case, the BU-NMF initialization relied on taking random values from the Gaussian distribution.
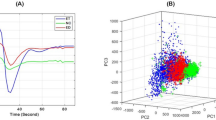
Random initialization of the BU-NMF algorithm was proven not to be repeatable as it gave ambiguous results when running it for multiple times [28]. For Patient #1, we ran the BU-NMF algorithm with two different random initializations and the results were completely different. We can see in Fig. 3 that BU-NMF in one case converged to a non-interpretable solution and only nine iterations were required (Fig. 3a, c). However, when the algorithm was initialized with different random values, it converged to interpretable results (Fig. 3b, d) and 376 iterations were required.
The number of iterations required for convergence of the BU-NMF algorithm, when ran for 50 times, is shown in Fig. 4 for all three patients. Fifty consecutive runs have been carried out by using the same ROI in each run, and different random values per voxel were produced each time for the algorithm initialization. The sets of points with the three different colors depicted in Fig. 4 correspond to each one of the three different patients.
For all three patients, the BU-NMF algorithm sometimes converged to interpretable solutions while others it did not. As explained before for Patient #1, when it converged to a non-interpretable solution, only a few iterations were required. On the other hand, when the algorithm converged to interpretable results, a greater number of iterations were required. Therefore, we concluded that the random initialization approach did not yield reproducible results and we opted for different initialization techniques, which are explained in the following paragraphs.
-
2.
Data-driven initialization: A number of such methods have also been tested, including a PK model as well as a simple gradient method. These initialization methods relied on information extracted directly from the data and when tested on the same patient data, similar results were obtained, which were also repeatable [28]. In particular, the wash-in and the ktrans maps were utilized for the initialization of the BU-NMF algorithm. In the first case, the BU-NMF algorithm was initialized by classifying the tumor area in three subareas using the “wash-in map” extracted from the first part of the time-signal curve related to each individual voxel. In the second initialization approach, the BU-NMF three-class initialization was derived by utilizing the ktrans map of ETM [26].
After computing the initial weight matrix W0, the H0 matrix was initialized by the least squares solution of A = W0*H0 where A is our given MR image data.
Concerning the decision on the value of k, for both random and data-driven initialization, PCA was applied to the data with the aim of determining the number of principal components that describe the 99% of the data variability. PCA is an orthogonal linear transformation, which projects the data onto a lower-dimensional subspace with respect to the original one. PCA involves the computation of the covariance matrix of the data and then its eigenvalues and corresponding eigenvectors. Eigenvectors representing the highest proportion of variance are the so-called principal components of the data. For the implementation of the PCA method, apart from the standard pre-processing reported previously, mean centering was also utilized, since this is a substantial requirement for PCA to work. After having identified the number of principal components, we used this information as an indication of how many different patterns the BU-NMF algorithm needs to seek in order to discover the underlying patterns of the MR data.
3 Results
For all three patients, an expert clinician has annotated the tumor region of interest within the entire 3D volume of the lesion. DCE-MRI data were acquired at 45 time points and for each patient, 384 × 384 × 14 pixels were used in our analysis. Data were organized in a two-dimensional data matrix X (i, t), where i is an index spanning all pixels in the 3D imaging space corresponding to the selected ROI and t denotes the signal time samples. In the next paragraphs, the results from the model-driven (PK) and model-free (PR) approach are presented along with a statistical comparison between the two methods. All figures depict results from a ROI selected by an expert clinician from the central slice of malignancy for each one of the patients.
3.1 Results from the pharmacokinetic model approach
ETM was applied to the data and in particular, the biomarkers ktrans and kep were estimated, the extracted maps of which are shown in Fig. 5b and c for the selected central slice (Fig. 5a) of Patient #1. The ktrans parameter is indicative of necrotic regions [25] and necrosis is associated with no enhancement regions having zero or close to zero ktrans values. On the other hand, well-perfused areas are those associated with high wash-in and wash-out values corresponding to high ktrans and kep accordingly. Last, it is important to mention that hypoxic regions are expected to have intermediate perfusion values, therefore it is hard to identify them in a straightforward way from the PK map.
To overcome the difficulty of approximating the hypoxic regions, three-region clustering of the ktrans map was performed using the k-means algorithm, resulting in a better distinction between the areas having different perfusion values. The ktrans and clustered ktrans maps depicted in Fig. 6a, b respectively, were obtained from Patient #1. Areas with high perfusion were assigned to green clusters, those with moderate perfusion to blue clusters and those with low or no perfusion to red clusters.
Initialization of the BU-NMF algorithm using the ktrans parameter of ETM for Patient #1. (a) The ktrans map. (b) The corresponding three-area clustering using k-means, with green clusters associated with areas of high perfusion, blue clusters with moderate perfusion areas and red ones with low or no perfusion areas
3.2 Results from the proposed model-free pattern recognition approach
As explained in Section 2, after the data pre-processing, PCA was applied to the data in order to discover the number of components that the BU-NMF algorithm will seek. For all three patients, we found that the total data variability was sufficiently (more than 99%) described by the three first three principal components identified by PCA.
The next step concerned the implementation of the BU-NMF algorithm. The algorithm was given as prior information: a) the number of patterns that are going to represent the DCE-MRI data, which was three as shown by the PCA results, and b) the initialized W0 and H0 matrices using the wash-in and PK initialization method, described previously in Section 2.
3.2.1 Initialization results
The first BU-NMF initialization approach relied on the extraction of the wash-in map, described in Section 2.3.2. A three-region classification map was subsequently derived from the wash-in map using the k-means algorithm. This map is depicted in Fig. 7a and it corresponds to Patient #2. Areas with high wash-in values are colored green, those with intermediate wash-in are blue and those with low or zero wash-in values are red.
Initialization of the BU-NMF algorithm using the wash-in and the PK map for Patient #2. (a) The three-region clustering map derived from k-means clustering on the wash-in map, with green clusters representing the high wash-in areas, blue representing the moderate wash-in areas and red ones representing the low or no wash-in areas. (b) The three-region clustering map derived from k-means clustering on the PK map, with green clusters representing the high perfusion areas, blue representing the moderate perfusion areas and red representing the low or no perfusion areas
The second BU-NMF initialization approach relied on the extraction of the ktrans map from the ETM model, similarly to the concept mentioned in the pharmacokinetic model method (Section 3.1). The extracted map from Patient #2 is illustrated in Fig. 7b.
The two initialization maps appear quite similar in terms of the necrotic region (red) but don’t seem to agree very much on the well-perfused (green) and hypoxic (blue) regions.
3.2.2 BU-NMF results
Results from both the wash-in and PK map initialization of the BU-NMF algorithm are illustrated in the following figures. In Figs. 8, 9, 10 the results from the BU-NMF implementation using wash-in initialization, are presented for Patients #1, #2 and #3 respectively. In Figs. 8a, 9a, 10a the plots of the three NMF components are depicted and in Figs. 8c, 9c, 10c the corresponding composite color maps are illustrated, showing the contribution of the three components to each individual voxel. Each image voxel is characterized by a mixture of well-perfused (green), hypoxic (blue) and necrotic (red) regions. The results of the PR analysis using the PK map initialization, containing the NMF plots and the composite color maps are shown in Figs. 8b, 9b, 10b and Figs. 8d, 9d, 10d respectively. We notice that for all three patients, the three different NMF components have the shape of the theoretically expected DCE curves as explained in Section 1, confirming the efficiency of the BU-NMF algorithm in automatically characterizing the enhancement profiles. In addition, considering the apparent high variability in the initial conditions the method in all cases yields consistent results. This is further investigated in the next section.
PR analysis results for Patient #1. Left: Plots of the three NMF components using the wash-in map (a) and the PK map initialization (b) respectively. Right: The corresponding composite color maps (c, d) derived from the two different initialization approaches describing the percentage contribution of the well-perfused (green), hypoxic (blue), and necrotic (red) components
PR analysis results for Patient #2. Left: Plots of the three NMF components using the wash-in map (a) and the PK map initialization (b) respectively. Right: The corresponding composite color maps (c, d) derived from the two different initialization approaches describing the percentage contribution of the well-perfused (green), hypoxic (blue), and necrotic (red) components
PR analysis results for Patient #3. Left: Plots of the three NMF components using the wash-in map (a) and the PK map initialization (b) respectively. Right: The corresponding composite color maps (c, d) derived from the two different initialization approaches describing the percentage contribution of the well-perfused (green), hypoxic (blue), and necrotic (red) components
3.3 Statistical analysis
Statistical analysis of our results was performed using Pearson’s linear correlation coefficient. The correlation coefficient can range between −1 and 1, which we corresponded to percentages ranging from −100% and 100%. A perfect positive correlation is represented by the value +1, the value 0 indicates no correlation and −1 indicates a perfect negative correlation. The closer the coefficient values are to 1 and −1, the stronger the relationship is between the variables. All image correlations described in the following paragraphs have been computed following image background removal, thus taking into account only the tumor ROI pixels.
In order to investigate the correspondence between the hypoxic, well-perfused and necrotic components derived from the two BU-NMF initialization approaches. The results are presented in Table 1 for all three patients.
We notice that the highest correlation percentages (greater than 91%) appear when the same components from the two different BU-NMF implementations are compared e.g. hypoxic component from wash-in initialization with hypoxic component from PK initialization. As expected, all comparisons of different components exhibit either low or negative correlations.
The relationship between the BU-NMF classification and the ktrans map three-region classification using k-means clustering, as explained in Section 3.1, was also investigated and the derived correlation percentages are presented in Table 2. The BU-NMF classification results were obtained from Patient #1 using the wash-in initialization method. Similar results were obtained when the PK initialization was chosen instead. Correlation is dominant when the same regions are compared even though percentages are not as high as in Table 1, ranging between 38.44% and 66.92%. The necrotic regions show the highest correlation (66.92%) whereas well-perfused and hypoxic components show lower correlation, i.e. 45.99% and 38.44% respectively. This might be due to the fact that the ktrans parameter is indicative of necrotic regions, as explained in Section 3.1, but is not able to distinguish well between the well-perfused and hypoxic regions when a simple classification method as k-means is used. Negative or low correlation percentages were observed when different components from the two methods were described.
In order to get a further understanding of the extracted tumor hypoxic regions, we focused on comparing only the hypoxic map extracted from the BU-NMF algorithm with the entire ktrans map (Table 3). Correlation percentages varied between 23.73–43.23%, which does not give very much evidence of correlation between the two methods.
Subsequently, we applied a double thresholding technique on the ktrans image in order to approximate the location of the hypoxic region in the tumor. The rationale for this is that the hypoxic areas will exhibit a ktrans profile greater than the necrotic (ktrans = 0) but less than the well-perfused areas (ktrans- > 1). We tested different threshold values and we identified the upper and lower bounds in the ktrans image that lead to the highest correlation between the image derived from ktrans thresholding and the hypoxic region derived from BU-NMF (Table 4). It is noticeable that for all three patients these bounds were between 0.15 and 0.76 approximately, with ktrans taking on values in a range of 0–1, leading to a maximum correlation with BU-NMF results of around 52%. From this, we could assume that the necrotic image regions according to our classification exhibit ktrans values lower than 0.15, well-perfused exhibit ktrans more than 0.76 and hypoxic regions lie in between these two thresholds. This assumption is reasonable since the hypoxic ktrans range is in line with findings claiming that the bulk of the tumor consists mainly of hypoxia; hence, the periphery of the tumor, which typically relates to the normoxic/well-perfused area, occupies smaller space [16, 25]. Necrotic regions instead are those with ktrans close to zero, occupying a smaller space with respect to hypoxia. However, in our case the necrotic region seemed to be overestimated by a small percentage of 15%, which can be explained by the limitation of MR imaging resolution regarding physiology, thus voxels with values close to zero that are adjacent to necrotic regions with ktrans = 0 are also considered as necrotic. Similar assumptions for the ktrans range were used in a recent study [19], which deals with the delineation of tumor physiological regions through the application of a cancer predictive model aiming to predict patients’ glioblastoma progress.
3.4 Qualitative findings from Histopathology reports
There is consistency in the morphological characterization of the three lesions regarding the existence of necrotic areas sporadically distributed within the tumors. In addition, all three tumors have been characterized by regionally high proliferation rates.
Specifically, for the patient with MPNST sarcoma, the immunohistochemical analysis has shown positive value of the proliferation marker (MIB-1), high cellularity as well as positive values of the microvessel density parameter CD34.
We can assume that localization of regions with high cellularity and vascularization, might indicate either lack of adequate oxygenation of the tumor, hence hypoxic regions, or presence of well-perfused areas.
4 Discussion
In the present work, three different approaches were tested on DCE-MRI data obtained from patients affected by sarcoma; the quantitative pharmacokinetic ETM model, the semi-quantitative BU-NMF algorithm applied on the time-signal DCE curves and the qualitative examination of the data provided by experienced radiologists. The ETM model provided us with the ktrans map, which is a well-established quantitative parameter for the localization of perfusion areas. As of the semi-quantitative PR approach, the BU-NMF algorithm was initialized by one random approach and two data-driven approaches. In terms of histopathological data, important tumor morphological information was obtained and for one of the three patients immunohistochemical information was also provided.
In particular, the three most common theoretically expected types of DCE tissue curve shapes (Types 1, 2 and 3) were confirmed by the results of the BU-NMF algorithm as being the dominant patterns that represent the signal. In addition, the well-perfused, hypoxic and necrotic components extracted from the two BU-NMF implementations (wash-in and PK initialized), correlated well between them while there was also a positive correlation with the same components extracted from the ktrans map with a simple k-means clustering. Small positive correlations were observed when comparing the ktrans image with the hypoxic component of BU-NMF. The correlations were significantly improved, i.e. improvement of 41% for wash-in and 48% for PK initialization, when only a constrained (via double thresholding) part of the ktrans map was compared against the hypoxic BU-NMF image. What is more, from the double thresholding technique we used on the ktrans image, the BU-NMF hypoxic regions best correlated to ktrans image with values between 0.15 and 0.76 approximately, well-perfused areas to ktrans above 0.76 and necrotic regions to ktrans below 0.15. These results are in agreement with published results and findings on tumor hypoxia [16, 25] as explained in Section 3.3. The histopathological findings indicated the presence of necrotic and well-perfused areas in all three lesions, however the detection of hypoxic regions would need further analysis since we only obtained immunohistochemistry results for one of the three patients.
In conclusion, the PR method used in this work gave fast (around 25 s execution time) and repeatable results, always converging after a precise number of iterations for each patient. In addition, the results obtained did not depend on complex fitting or initialization procedures in contrast to computationally expensive PK models, which also require non-linear fitting schemes that may lead to local extrema problems. After successfully addressing the problems related to random initialization, the presented method was robust to initial conditions since it gave very similar results for both initialization schemes. In addition, user interaction was minimal, whereas for PK models there is need to select certain parameters in advance such as the AIF [3]. Last but not least, in contrast to model-based approaches, our method does not make any assumptions on the underlying biology since it is purely data-driven.
The findings stemming from the present study may be useful in many different ways. Tumor classification based on hypoxia, could be very helpful in determining the malignancy grade of a tumor and most importantly in assisting the oncologist to suggest more effective treatment planning and possibly limit or even avoid the radiation therapy. Achieving hypoxia delineation and mapping of the diverse perfusion areas present in heterogeneous tumors, will be also useful in guiding core needle biopsy so that it targets the most representative areas of the tumor. In addition, it could help to differentiate tumors based on the perfusion maps extracted from the MRI examination that could play a key role in deciding whether surgery is needed or not.
An obvious limitation of the current work is further validation against histopathology and immunohistochemistry, which was not available for all three sarcomas. To this end, the current work focused mainly on demonstrating the increased stability and invariance to initial conditions that previous work suffered [27] and on correlating the automatically extracted regions to the well-known ktrans variable. Interestingly, our results show that by containing the ktrans parametrized image with a double threshold (lower to exclude necrosis and upper to exclude well-perfused areas), the correlation to our extracted hypoxic components increases significantly. This is an encouraging indication that our data-based approach is able to produce clinically significant results related to tumor hypoxia, a hypothesis that needs further investigation in order to be confirmed.
To this end, we will also compare a large number of pharmacokinetic parameters from a wide range of DCE-MRI models such as Tofts and GCTT [20], in order to examine possible correlations more thoroughly and compare them to already published work from studies investigating the relationship of DCE-MRI to tumor hypoxia. The main plan for future work is to compare our computational results from the model-free and model-based approach with quantitative histopathological parameters. All the tissue sections analyzed computationally, will be histopathologically examined for morphological features, while additional immunohistochemistry will identify biomarkers, which will be afterwards translated to hypoxia and other related cancer hallmarks. What is more, further work and analysis on a large series of tumor image data will be performed in order to confirm our results and investigate the correlations between model-based, model-free methods and histopathology in different tumor malignancies.
5 Conclusion
The present study analyzed the tumor physiology of three histologically different sarcomas by using non-invasive DCE-MR imaging data, which were post-processed leading to quantitative and semi-quantitative results. An advanced matrix factorization algorithm, BU-NMF, has been applied to the data and its results were compared to the pharmacokinetic ETM model and to histopathology findings as well. Our results indicate that hypoxia can be estimated from non-invasive DCE-MR imaging data using purely data-driven methods.
The BU-NMF algorithm was proved to be robust, as it gave consistent results when initialized by different methods. It yielded to the identification of the three most common enhancement patterns of the DCE time-signal uptake curves, which is in line with theoretical findings claiming that there are three types of DCE tissue curves as explained in Section 1. BU-NMF also segmented the tumor area in three regions characterized by different perfusion, i.e. well-perfused, hypoxic and necrotic one, which correlated well when compared with the same areas extracted from the ktrans parametric map of the pharmacokinetic ETM model. Interestingly, the BU-NMF hypoxic map seems to correlate with a constrained ktrans image, as is theoretically expected. What is more, from the histopathological findings, the existence of necrotic and well-perfused areas in the three lesions was further confirmed.
The current study enabled us to obtain an objective evaluation of the physiology of the lesions under examination, which was supported by the qualitative analysis of the experts. Once this is validated, it could be particularly useful as an imaging hypoxia biomarker for monitoring the follow up of the patient. To this end, additional studies have been planned to test the algorithm on larger patient cohorts and to validate the results through histopathology, with the ultimate goal to develop an automated tool for the detection of hypoxia using DCE-MRI.
Change history
05 January 2018
The article Pattern recognition and pharmacokinetic methods on DCE-MRI data for tumor hypoxia mapping in sarcoma, written by M. Venianaki, O. Salvetti, E. de Bree, T. Maris, A. Karantanas, E. Kontopodis, K. Nikiforaki, K. Marias, was originally published electronically without open access.
References
Berry MW, Browne M, Langville AN, Pauca VP, Plemmons RJ (2007) Algorithms and applications for approximate nonnegative matrix factorization. Comput Stat Data Anal 52(1):155–173. https://doi.org/10.1016/j.csda.2006.11.006
Cho H, Ackerstaff E, Carlin S et al (2009) Noninvasive Multimodality Imaging of the Tumor Microenvironment: Registered Dynamic Magnetic Resonance Imaging and Positron Emission Tomography Studies of a Preclinical Tumor Model of Tumor Hypoxia. Neoplasia 11(3):247IN2–259IN3. https://doi.org/10.1593/neo.81360
Eyal E, Degani H (2009) Model-based and model-free parametric analysis of breast dynamic-contrast-enhanced MRI. NMR Biomed 22(1):40–53. https://doi.org/10.1002/nbm.1221
Fisher SM, Joodi R, Madhuranthakam AJ, Öz OK, Sharma R, Chhabra A (2016) Current utilities of imaging in grading musculoskeletal soft tissue sarcomas. Eur J Radiol 85(7):1336–1344. https://doi.org/10.1016/j.ejrad.2016.05.003
Fukumura D, Jain RK (2007) Tumor microenvironment abnormalities: Causes, consequences, and strategies to normalize. J Cell Biochem 101(4):937–949. https://doi.org/10.1002/jcb.21187
Han SH, Ackerstaff E, Stoyanova R et al (2013) Gaussian mixture model-based classification of dynamic contrast enhanced MRI data for identifying diverse tumor microenvironments: preliminary results. NMR Biomed 26(5):519–532. https://doi.org/10.1002/nbm.2888
Höckel M, Schlenger K, Mitze M, Schäffer U, Vaupel P (1996) Hypoxia and radiation response in human tumors. Semin Radiat Oncol 6(1):3–9. https://doi.org/10.1016/S1053-4296(96)80031-2
Hoffmann U, Brix G, Knopp MV, Heβ T, Lorenz WJ (1995) Pharmacokinetic Mapping of the Breast: A New Method for Dynamic MR Mammography. Magn Reson Med 33(4):506–514. https://doi.org/10.1002/mrm.1910330408
Jensen RL, Mumert ML, Gillespie DL, Kinney AY, Schabel MC, Salzman KL (2014) Preoperative dynamic contrast-enhanced MRI correlates with molecular markers of hypoxia and vascularity in specific areas of intratumoral microenvironment and is predictive of patient outcome. Neuro Oncol 16(2):280–291. https://doi.org/10.1093/neuonc/not148
Knopp MV, Weiss E, Sinn HP et al (1999) Pathophysiologic basis of contrast enhancement in breast tumors. J Magn Reson Imaging 10(3):260–266. https://doi.org/10.1002/(SICI)1522-2586(199909)10:3<260::AID-JMRI6>3.0.CO;2-7
Kontopodis E, Karatzanis I, Sakkalis V, Francesca B, Marias K (2016) A DCE-MRI analysis workflow. CGI ’16:Proceedings of the 33rd Computer Graphics International conference, 101–104. https://doi.org/10.1145/2949035.2949061
Kuhl CK, Mielcareck P, Klaschik S et al (1999) Dynamic breast MR imaging: are signal intensity time course data useful for differential diagnosis of enhancing lesions? Radiology 211:101–110. https://doi.org/10.1148/radiology.211.1.r99ap38101
Lee DD, Seung HS (1999) Learning the parts of objects by non-negative matrix factorization. Nature 401(6755):788–791. https://doi.org/10.1038/44565
Lee DD, Seung HS (2001) Algorithms for Non-negative Matrix Factorization. Adv Neural Inf Proces Syst 13:556–562
Menon C, Fraker DL (2005) Tumor oxygenation status as a prognostic marker. Cancer Lett 221(2):225–235. https://doi.org/10.1016/j.canlet.2004.06.029
Neal ML, Trister AD, Cloke T et al (2013) Discriminating Survival Outcomes in Patients with Glioblastoma Using a Simulation-Based, Patient-Specific Response Metric. PLoS One 8(1):e51951. https://doi.org/10.1371/journal.pone.0051951
Newbold K, Castellano I, Charles-Edwards E et al (2009) An Exploratory Study Into the Role of Dynamic Contrast-Enhanced Magnetic Resonance Imaging or Perfusion Computed Tomography for Detection of Intratumoral Hypoxia in Head-and-Neck Cancer. Int J Radiat Oncol Biol Phys 74(1):29–37. https://doi.org/10.1016/j.ijrobp.2008.07.039
Paatero P, Tapper U (1994) Positive matrix factorization: A non-negative factor model with optimal utilization of error estimates of data values. Environmetrics 5(2):111–126. https://doi.org/10.1002/env.3170050203
Roniotis A, Oraiopoulou M-E, Tzamali E et al (2015) A Proposed Paradigm Shift in Initializing Cancer Predictive Models with DCE-MRI Based PK Parameters: A Feasibility Study. Cancer Informat 14(Suppl 4):7. https://doi.org/10.4137/CIN.S19339
Schabel MC (2012) A unified impulse response model for DCE-MRI. Magn Reson Med 68(5):1632–1646. https://doi.org/10.1002/mrm.24162
Soldatos T, Ahlawat S, Montgomery E, Chalian M, Jacobs MA, Fayad LM (2016) Multiparametric Assessment of Treatment Response in High-Grade Soft-Tissue Sarcomas with Anatomic and Functional MR Imaging Sequences. Radiology 278(3):831–840. https://doi.org/10.1148/radiol.2015142463
Sourbron SP, Buckley DL (2011) On the scope and interpretation of the Tofts models for DCE-MRI. Magn Reson Med 66(3):735–745. https://doi.org/10.1002/mrm.22861
Stoyanova R, Huang K, Sandler K et al (2012) Mapping Tumor Hypoxia In Vivo Using Pattern Recognition of Dynamic Contrast-enhanced MRI Data. Transl Oncol 5(6):437–IN2. https://doi.org/10.1593/tlo.12319
Surov A, Meyer HJ, Gawlitza M et al (2017) Correlations Between DCE MRI and Histopathological Parameters in Head and Neck Squamous Cell Carcinoma. Transl Oncol 10(1):17–21. https://doi.org/10.1016/j.tranon. 2016.10.001
Swanson KR, Chakraborty G, Wang CH et al (2009) Complementary but distinct roles for MRI and 18F-fluoromisonidazole PET in the assessment of human glioblastomas. J Nucl Med 50(1):36–44. https://doi.org/10.2967/jnumed.108.055467
Tofts PS, Kermode AG (1991) Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn Reson Med 17(2):357–367. https://doi.org/10.1002/mrm.1910170208
Venianaki M, Kontopodis E, Nikiforaki K, de Bree E, Salvetti O, Marias K (2016) A model-free approach for imaging tumor hypoxia from DCE-MRI data. CGI ’16:Proceedings of the 33rd Computer Graphics International conference, 105–108. https://doi.org/10.1145/2949035.2949062
Venianaki M, Kontopodis E, Nikiforaki K, et al (2016) Improving hypoxia map estimation by using model-free classification techniques in DCE-MRI images. 2016 IEEE International Conference on Imaging Systems and Techniques (IST), 183–188. https://doi.org/10.1109/IST.2016.7738220
Walsh JC, Lebedev A, Aten E, Madsen K, Marciano L, Kolb HC (2014) The clinical importance of assessing tumor hypoxia: relationship of tumor hypoxia to prognosis and therapeutic opportunities. Antioxid Redox Signal 21(10):1516–1554. https://doi.org/10.1089/ars.2013.5378
Xu Y, Yin W (2013) A block coordinate descent method for regularized multiconvex optimization with applications to nonnegative tensor factorization and completion. SIAM J Imag Sci 6(3):1758–1789. https://doi.org/10.1137/120887795
Zheng L, Li Y, Geng F et al (2015) Using semi-quantitative dynamic contrast-enhanced magnetic resonance imaging parameters to evaluate tumor hypoxia: a preclinical feasibility study in a maxillofacial VX2 rabbit model. Am J Transl Res 7(3):535–547
Acknowledgments
KM acknowledges support from the CHIC project GA600841 funded by the 7th Framework Programme of the European Commission. The authors thank Mariam-Eleni Oraiopoulou for helpful discussions and comments regarding tumor physiology that greatly improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
The original version of this article was revised due to a retrospective Open Access order.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Venianaki, M., Salvetti, O., de Bree, E. et al. Pattern recognition and pharmacokinetic methods on DCE-MRI data for tumor hypoxia mapping in sarcoma. Multimed Tools Appl 77, 9417–9439 (2018). https://doi.org/10.1007/s11042-017-5046-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-5046-6