Abstract
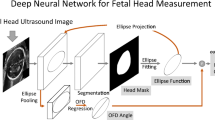
Accurately segmenting the structure of the fetal head (FH) and performing biometry measurements, including head circumference (HC) estimation, stands as a vital requirement for addressing abnormal fetal growth during pregnancy under the expertise of experienced radiologists using ultrasound (US) images. However, accurate segmentation and measurement is a challenging task due to image artifact, incomplete ellipse fitting, and fluctuations due to FH dimensions over different trimesters. Also, it is highly time-consuming due to the absence of specialized features, which leads to low segmentation accuracy. To address these challenging tasks, we propose an automatic density regression approach to incorporate appearance and shape priors into the deep learning-based network model (DR-ASPnet) with robust ellipse fitting using fetal US images. Initially, we employed multiple pre-processing steps to remove unwanted distortions, variable fluctuations, and a clear view of significant features from the US images. Then some form of augmentation operation is applied to increase the diversity of the dataset. Next, we proposed the hierarchical density regression deep convolutional neural network (HDR-DCNN) model, which involves three network models to determine the complex location of FH for accurate segmentation during the training and testing processes. Then, we used post-processing operations using contrast enhancement filtering with a morphological operation model to smooth the region and remove unnecessary artifacts from the segmentation results. After post-processing, we applied the smoothed segmented result to the robust ellipse fitting-based least square (REFLS) method for HC estimation. Experimental results of the DR-ASPnet model obtain 98.86% dice similarity coefficient (DSC) as segmentation accuracy, and it also obtains 1.67 mm absolute distance (AD) as measurement accuracy compared to other state-of-the-art methods. Finally, we achieved a 0.99 correlation coefficient (CC) in estimating the measured and predicted HC values on the HC18 dataset.












Similar content being viewed by others
Data Availability
The dataset is public and can be downloaded from https://hc18.grand-challenge.org/.
References
Figueras F, Caradeux J, Crispi F, Eixarch E, Peguero A, GratacosE: Diagnosis and surveillance of late-onset fetal growth restriction. American journal of obstetrics and gynecology. 218(2):S790-802, 2018
Necas M: The clinical ultrasound report: Guideline for sonographers. Australasian Journal of Ultrasound in Medicine. 21(1):9-23, 2018
Matthew J, Skelton E, Day TG, Zimmer VA, Gomez A, Wheeler G, Toussaint N, Liu T, Budd S, Lloyd K, Wright R: Exploring a new paradigm for the fetal anomaly ultrasound scan: Artificial intelligence in real time. Prenatal diagnosis. 42(1):49-59, 2022
World Health Organization. WHO antenatal care recommendations for a positive pregnancy experience: maternal and fetal assessment update: imaging ultrasound before 24 weeks of pregnancy.
Mamsen LS, Björvang RD, Mucs D, Vinnars MT, Papadogiannakis N, Lindh CH, Andersen CY, Damdimopoulou P: Concentrations of perfluoroalkyl substances (PFASs) in human embryonic and fetal organs from first, second, and third trimester pregnancies. Environment international. 124:482-92, 2019
O'Gorman N, Salomon LJ: Fetal biometry to assess the size and growth of the fetus. Best practice & research Clinical obstetrics & gynaecology. 49:3-15, 2018
Mandal SK, Ghosh SK, Roy S, Prakash B: Evaluation of fetaltranscerebellar diameter as a sonological parameter for the estimation of fetal gestational age in comparison to biparietal diameter and femur length. age. 6:7, 2019
Hammami A, Mazer Zumaeta A, Syngelaki A, Akolekar R, Nicolaides KH: Ultrasonographic estimation of fetal weight: development of new model and assessment of performance of previous models. Ultrasound in obstetrics &gynecology. 52(1):35-43, 2018
O'Brien CM, Louise J, Deussen A, Dodd JM: In overweight and obese women, fetal ultrasound biometry accurately predicts newborn measures. Australian and New Zealand Journal of Obstetrics and Gynaecology. 60(1):101-7, 2020
Salomon LJ, Alfirevic Z, Da Silva Costa F, Deter RL, Figueras F, Ghi TA, Glanc P, Khalil A, Lee W, Napolitano R, Papageorghiou A: ISUOG Practice Guidelines: ultrasound assessment of fetal biometry and growth. Ultrasound in obstetrics &gynecology. 53(6):715-23, 2019
Burgos-Artizzu XP, Coronado-Gutiérrez D, Valenzuela-Alcaraz B, Vellvé K, Eixarch E, Crispi F, Bonet-Carne E, Bennasar M, Gratacos E: Analysis of maturation features in fetal brain ultrasound via artificial intelligence for the estimation of gestational age. American Journal of Obstetrics &Gynecology MFM. 3(6):100462, 2021
Sun Y, Yang H, Zhou J, Wang Y: ISSMF: Integrated semantic and spatial information of multi-level features for automatic segmentation in prenatal ultrasound images. Artificial Intelligence in Medicine. 125:102254, 2022
AdithyaPC, Sankar R, Moreno WA, Hart S: Trends in fetal monitoring through phonocardiography: Challenges and future directions. Biomedical Signal Processing and Control. 33:289–305, 2017
Kiserud T, Piaggio G, Carroli G, Widmer M, Carvalho J, Neerup Jensen L, Giordano D, Cecatti JG, Abdel Aleem H,Talegawkar SA, Benachi A: The World Health Organization fetal growth charts: a multinational longitudinal study of ultrasound biometric measurements and estimated fetal weight. PLoS medicine. 14(1):e1002220, 2017
Sahli H, Ben Slama A, MouelhiA, Soayeh N, Rachdi R, Sayadi M: A computer-aided method based on geometrical texture features for a precocious detection of fetal Hydrocephalus in ultrasound images. Technology and Health Care. 28(6):643-64, 2020
Rajinikanth V, Dey N, Kumar R,Panneerselvam J, Raja NS: Fetal head periphery extraction from ultrasound image using Jaya algorithm and Chan-Vese segmentation. Procedia Computer Science. 152:66-73, 2019
Zhang L, Dudley NJ, Lambrou T, Allinson N, Ye X: Automatic image quality assessment and measurement of fetal head in two-dimensional ultrasound image. Journal of Medical Imaging. 4(2):024001, 2017
Cerrolaza JJ, Oktay O, Gomez A, Matthew J, Knight C, Kainz B, Rueckert D: Fetal skull segmentation in 3D ultrasound via structured geodesic random forest. In Fetal, Infant and Ophthalmic Medical Image Analysis: International Workshop, FIFI 2017, and 4th International Workshop, OMIA 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings 4 (pp. 25–32). Springer International Publishing, 2017
Komatsu M, Sakai A, Dozen A, Shozu K, Yasutomi S, Machino H, Asada K, Kaneko S, Hamamoto R: Towards clinical application of artificial intelligence in ultrasound imaging. Biomedicines. 9(7):720, 2021
Cerrolaza JJ, Sinclair M, Li Y, Gomez A, Ferrante E, Matthew J, Gupta C, Knight CL, Rueckert D: Deep learning with ultrasound physics for fetal skull segmentation. In2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) (pp. 564–567). IEEE, 2018
Skeika EL, Da Luz MR, Fernandes BJ, Siqueira HV, De Andrade ML: Convolutional neural network to detect and measure fetal skull circumference in ultrasound imaging. IEEE Access. 8:191519-29, 2020
Sobhaninia Z, Emami A, Karimi N, Samavi S: Localization of fetal head in ultrasound images by multiscale view and deep neural networks. In2020 25th International Computer Conference, Computer Society of Iran (CSICC) (pp. 1–5). IEEE, 2020
Wu L, Xin Y, Li S, Wang T, Heng PA, Ni D: Cascaded fully convolutional networks for automatic prenatal ultrasound image segmentation. In2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017) (pp. 663–666). IEEE, 2017
Qiao D, Zulkernine F: Dilated squeeze-and-excitation U-Net for fetal ultrasound image segmentation. In2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB) (pp. 1–7). IEEE, 2020
Zhang J,Petitjean C, Lopez P, Ainouz S: Direct estimation of fetal head circumference from ultrasound images based on regression CNN. InMedical Imaging with Deep Learning (pp. 914–922). PMLR, 2020
Zhao L, Li N, Tan G, Chen J, Li S, Duan M: The End-to-end Fetal Head Circumference Detection and Estimation in Ultrasound Images. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022
Li J, Wang Y, Lei B, Cheng JZ, Qin J, Wang T, Li S, Ni D: Automatic FH circumference measurement in ultrasound using random forest and fast ellipse fitting. IEEE journal of biomedical and health informatics. 22(1):215-23, 2017
van den Heuvel TL, de Bruijn D, de Korte CL, Ginneken BV: Automated measurement of fetal head circumference using 2D ultrasound images. PloS one. 13(8):e0200412, 2018
Fiorentino MC, Moccia S, Capparuccini M, Giamberini S, Frontoni E: A regression framework to head-circumference delineation from US fetal images. Computer methods and programs in biomedicine. 198:105771, 2021
Zeng Y, Tsui PH, Wu W, Zhou Z, Wu S: Fetal ultrasound image segmentation for automatic head circumference biometry using deeply supervised attention-gated V-Net. Journal of Digital Imaging. 34:134-48, 2021
Amini SM: Head circumference measurement with deep learning approach based on multi-scale ultrasound images. Multimedia Tools and Applications. 81(23):32981-93, 2022
Zhou M, Wang C, Lu Y, Qiu R, Zeng R, Zhi D, Jiang X, Ou Z, Wang H, Chen G, Bai J: The segmentation effect of style transfer on FH ultrasound image: a study of multi-source data. Medical & Biological Engineering & Computing. 1–5, 2023
Pazinato DV, Stein BV, de Almeida WR, Werneck RD, Júnior PR, Penatti OA, Torres RD, Menezes FH, Rocha A: Pixel-level tissue classification for ultrasound images. IEEE journal of biomedical and health informatics. 20(1):256-67, 2014
Medak D, Posilović L, Subašić M, Budimir M, Lončarić S: Automated defect detection from ultrasonic images using deep learning. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. 68(10):3126-34, 2021
Korneev S, Narodytska N, Pulina L, Tacchella A, Bjorner N, Sagiv M: Constrained image generation using binarized neural networks with decision procedures. InTheory and Applications of Satisfiability Testing–SAT 2018: 21st International Conference, SAT 2018, Held as Part of the Federated Logic Conference, FloC 2018, Oxford, UK, July 9–12, 2018, Proceedings 21 (pp. 438–449). Springer International Publishing, 2018
Wu L, Cheng JZ, Li S, Lei B, Wang T, Ni D: FUIQA: fetal ultrasound image quality assessment with deep convolutional networks. IEEE transactions on cybernetics. 47(5):1336-49, 2017
Kulkarni M, Karande S: Layer-wise training of deep networks using kernel similarity. arXiv preprint http://arxiv.org/abs/1703.07115, 2017
Sun Y, Xue B, Zhang M, Yen GG: Completely automated CNN architecture design based on blocks. IEEE transactions on neural networks and learning systems. 31(4):1242-54, 2019
Liu Y, Gao Y, Yin W: An improved analysis of stochastic gradient descent with momentum. Advances in Neural Information Processing Systems. 33:18261-71, 2020
Bustacara-Medina C, Flórez-Valencia L:An automatic stopping criterion for contrast enhancement using multi-scale top-hat transformation, Sens. Imaging. 20(1):26, 2019
Liang J, Zhang M, Liu D, Zeng X, Ojowu O, Zhao K, Li Z, Liu H: Robust ellipse fitting based on sparse combination of data points. IEEE Trans. Image Process. 22(6):2207–2218, 2013
Dice LR: Measures of the amount of ecologic association between species. Ecology. 26(3):297–302, 1945
Babalola KO, Patenaude B, Aljabar P, Schnabel J, Kennedy D, Crum W, et al: Comparison and Evaluation of Segmentation Techniques for Subcortical Structures in Brain MRI. Med Image ComputComputInterv – MICCAI 2008, Springer, Berlin, Heidelberg, p. 409–16, 2008
Shozu K, Komatsu M, Sakai A, Komatsu R, Dozen A, Machino H, Yasutomi S, Arakaki T, Asada K, Kaneko S, Matsuoka R: Model-agnostic method for thoracic wall segmentation in fetal ultrasound videos. Biomolecules. 10(12):1691, 2020
Roelfsema NM, Hop WC, Boito SM, Wladimiroff JW: Three-dimensional sonographic measurement of normal fetal brain volume during the second half of pregnancy. American journal of obstetrics and gynecology. 190(1):275-80, 2004
Sinclair M, Baumgartner CF, Matthew J, Bai W, Martinez JC, Li Y, Smith S, Knight CL, Kainz B, Hajnal J, King AP: Human-level performance on automatic head biometrics in fetal ultrasound using fully convolutional neural networks. In2018 40th annual international conference of the IEEE engineering in medicine and biology society (EMBC) (pp. 714–717). IEEE, 2018
Al-Bander B, Alzahrani T, Alzahrani S, Williams BM, Zheng Y:Improving fetal head contour detection by object localisation with deep learning, Annual Conference on Medical Image Understanding and Analysis, Springer, pp. 142–150, 2019
Liu P, Zhao H, Li P, Cao F: Automated classification and measurement of fetal ultrasound images with attention feature pyramid network, Second Target Recognition and Artificial Intelligence Summit Forum, SPIE, p. 114272R, 2020
Li P, Zhao H, Liu P, Cao F: Automated measurement network for accurate segmentation and parameter modification in fetal head ultrasound images. Medical & Biological Engineering & Computing. 58:2879-92, 2020
Moccia S, Fiorentino MC, Frontoni E: Mask-R2 CNN: A distance-field regression version of Mask-RCNN for fetal-head delineation in ultrasound images. Int. J. Comput. Assist. Radiol. Surg. 16:1711–1718, 2021
Alzubaidi M, Agus M, Shah U, Makhlouf M, Alyafei K, Househ M: Ensemble Transfer Learning for Fetal Head Analysis: From Segmentation to Gestational Age and Weight Prediction. Diagnostics. 12(9):2229, 2022
Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, Wang Q, Shen D: Medical image synthesis with deep convolutional adversarial networks. IEEE Transactions on Biomedical Engineering. 65(12):2720-30, 2018
Wang G, Li W, Aertsen M, Deprest J, Ourselin S, Vercauteren T. Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks. Neurocomputing. 338:34-45, 2019
Author information
Authors and Affiliations
Contributions
All the authors have participated in writing the manuscript and have revised the final version. All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by GD, SS, AKJ, MS, PS, and MM. The first draft of the manuscript was written by GD, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. Conceptualization: GD, SS. Methodology: GD, AKJ. Formal analysis and investigation: GD, MS, SS. Writing — original draft preparation: GD, PS, MM. Writing — review and editing: GD, AKJ, MS. Supervision: MM.
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants and/or animals performed by any of the authors.
Informed Consent
There is no informed consent for this study.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dubey, G., Srivastava, S., Jayswal, A.K. et al. Fetal Ultrasound Segmentation and Measurements Using Appearance and Shape Prior Based Density Regression with Deep CNN and Robust Ellipse Fitting. J Digit Imaging. Inform. med. 37, 247–267 (2024). https://doi.org/10.1007/s10278-023-00908-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-023-00908-8