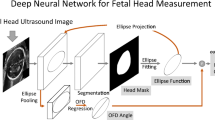
Abstract
Automatic computerized segmentation of fetal head from ultrasound images and head circumference (HC) biometric measurement is still challenging, due to the inherent characteristics of fetal ultrasound images at different semesters of pregnancy. In this paper, we proposed a new deep learning method for automatic fetal ultrasound image segmentation and HC biometry: deeply supervised attention-gated (DAG) V-Net, which incorporated the attention mechanism and deep supervision strategy into V-Net models. In addition, multi-scale loss function was introduced for deep supervision. The training set of the HC18 Challenge was expanded with data augmentation to train the DAG V-Net deep learning models. The trained models were used to automatically segment fetal head from two-dimensional ultrasound images, followed by morphological processing, edge detection, and ellipse fitting. The fitted ellipses were then used for HC biometric measurement. The proposed DAG V-Net method was evaluated on the testing set of HC18 (n = 355), in terms of four performance indices: Dice similarity coefficient (DSC), Hausdorff distance (HD), HC difference (DF), and HC absolute difference (ADF). Experimental results showed that DAG V-Net had a DSC of 97.93%, a DF of 0.09 ± 2.45 mm, an AD of 1.77 ± 1.69 mm, and an HD of 1.29 ± 0.79 mm. The proposed DAG V-Net method ranks fifth among the participants in the HC18 Challenge. By incorporating the attention mechanism and deep supervision, the proposed method yielded better segmentation performance than conventional U-Net and V-Net methods. Compared with published state-of-the-art methods, the proposed DAG V-Net had better or comparable segmentation performance. The proposed DAG V-Net may be used as a new method for fetal ultrasound image segmentation and HC biometry. The code of DAG V-Net will be made available publicly on https://github.com/xiaojinmao-code/.










Similar content being viewed by others
References
Sobhaninia Z, Emami A, Karimi N, Samavi S: Localization of fetal head in ultrasound images by multiscale view and deep neural networks. arXiv preprint, 2019 https://arxiv.org/abs/1911.00908
Jatmiko W, Habibie I, Ma'sum MA, Rahmatullah R, Satwika IP: Automated telehealth system for fetal growth detection and approximation of ultrasound images. Int J Smart Sensing Intell Syst 8(1):697-719,2015
Schmidt U, Temerinac D, Bildstein K, Tuschy B, Mayer J, Sütterlin M, Siemer J, Kehl S: Finding the most accurate method to measure head circumference for fetal weight estimation. Eur J Obstet Gynecol Reprod Biol 178:153-156,2014
Rueda S, Fathima S, Knight CL, Yaqub M, Papageorghiou AT, Rahmatullah B, Foi A, Maggioni M, Pepe A, Tohka J, Stebbing RV, McManigle JE, Ciurte A, Bresson X, Cuadra MB, Sun C, Ponomarev GV, Gelfand M.S, Kazanov MD, Wang CW, Chen HC, Peng CW, Hung CM, Noble JA: Evaluation and comparison of current fetal ultrasound image segmentation methods for biometric measurements: A grand challenge, IEEE Trans Med Imaging 33(4):797-813,2013
Jardim SMGVB, Figueiredo MAT: Segmentation of fetal ultrasound images. Ultrasound Med Biol 31(2):243–250,2005
Perez-Gonzalez JL, Muńoz JCB, Porras MCR, Arámbula-Cosío F, Medina-Bańuelos V: Automatic fetal head measurements from ultrasound images using optimal ellipse detection and texture maps, VI Latin American Congress on Biomedical Engineering CLAIB 2014, Springer, 2015, pp. 329-332
Ponomarev GV, Gelfand MS, Kazanov MD: A multilevel thresholding combined with edge detection and shape-based recognition for segmentation of fetal ultrasound images, Proceedings of Challenge US: Biometric Measurements from Fetal Ultrasound Images, ISBI 2012, 2012, pp. 17-19
Shrimali V, Anand R, Kumar V: Improved segmentation of ultrasound images for fetal biometry, using morphological operators, 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE, 2009, pp. 459-462
Lu W, Tan J, Floyd R: And biology, Automated fetal head detection and measurement in ultrasound images by iterative randomized Hough transform, Ultrasound Med. Biol. 31(7):929–936,2005
Zhang L, Ye X, Lambrou T, Duan W, Allinson N, Dudley NJ: A supervised texton based approach for automatic segmentation and measurement of the fetal head and femur in 2D ultrasound images. Phys Med Biol 61(3):1095–1115,2016
Li J, Wang Y, Lei B, Cheng JZ, Qin J, Wang T, Li S, Ni D: Automatic fetal head circumference measurement in ultrasound using random forest and fast ellipse fitting. IEEE J Biomed Health Inform 22(1):215-223,2017
van den Heuvel TL, de Bruijn D, de Korte CL, van Ginneken B: Automated measurement of fetal head circumference using 2D ultrasound images. PLoS One 13(8):e0200412,2018
Long J, Shelhamer E, Darrell T: Fully convolutional networks for semantic segmentation, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, IEEE, 2015, pp. 3431–3440
Ronneberger O, Fischer P, Brox T: U-net: Convolutional networks for biomedical image segmentation, International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, 2015, pp. 234–241
Milletari F, Navab N, Ahmadi SA: V-net: Fully convolutional neural networks for volumetric medical image segmentation, 2016 Fourth International Conference on 3D Vision (3DV), IEEE, 2016, pp. 565–571
Jang J, Park Y, Kim B, Lee SM, Kwon JY, Seo JK: Automatic estimation of fetal abdominal circumference from ultrasound images. IEEE J Biomed Health Inform 22(5):1512-1520,2017
Wu L, Xin Y, Li S, Wang T, Heng PA, Ni D: Cascaded fully convolutional networks for automatic prenatal ultrasound image segmentation, 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), IEEE, 2017, pp. 663-666
Al-Bander B, Alzahrani T, Alzahrani S, Williams BM, Zheng Y: Improving fetal head contour detection by object localisation with deep learning, Annual Conference on Medical Image Understanding and Analysis, Springer, 2019, pp. 142–150
Dou Q, Yu LH, Chen H, Jin Y, Yang X, Qin J, Heng PA: 3D deeply supervised network for automated segmentation of volumetric medical images, Med. Image Anal. 41, 2017, 40-54
Wu Y, He K: Group normalization, Proceedings of the European Conference on Computer Vision (ECCV), Springer, 2018, pp. 3–19
Ioffe S, Szegedy C: Batch normalization: Accelerating deep network training by reducing internal covariate shift, arXiv preprint, 2015 https://arxiv.org/abs/1502.03167
Springenberg JT, Dosovitskiy A, Brox T, Riedmiller M: Striving for simplicity: the all convolutional net, arXiv preprint, 2014 https://arxiv.org/abs/1412.6806
Zhu Q, Du B, Turkbey B, Choyke PL, Yan P: Deeply-supervised CNN for prostate segmentation, 2017 International Joint Conference on Neural Networks, IEEE, 2017, pp. 178–184
Shen T, Zhou T, Long G, Jiang J, Pan S, Zhang C: DiSAN: Directional self-attention network for RNN/CNN-free language understanding, Thirty-Second AAAI Conference on Artificial Intelligence, AAAI Publications, 2018, pp. 5446–5455
Roth HR, Lu L, Lay N, Harrison AP, Farag A, Sohn A, Summers RM: Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation, Med. Image Anal. 45, 2018, 94-107
Roth HR, Oda H, Hayashi Y, Oda M, Shimizu N, Fujiwara M, Misawa KK Mori K: Hierarchical 3D fully convolutional networks for multi-organ segmentation, arXiv preprint, 2017 https://arxiv.org/abs/1704.06382
Schlemper J, Oktay O, Chen L, Matthew J, Knight C, Kainz B, Glocker B, Rueckert D: Attention-gated networks for improving ultrasound scan plane detection, arXiv preprint, 2018 https://arxiv.org/abs/1804.05338
Zhang S, Fu H, Yan Y, Zhang Y, Wu Q, Yang M, Tan M, Xu Y: Attention guided network for retinal image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, 2019, pp. 797–805
Wang X, Girshick R, Gupta A, He K: Non-local neural networks, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, IEEE, 2018, pp. 7794–7803
Kingma DP, Ba J: Adam: A method for stochastic optimization, arXiv preprint, 2014 https://arxiv.org/abs/1412.6980
Rong Y, Xiang D, Zhu W, Shi F, Gao E, Fan Z, Chen X: Deriving external forces via convolutional neural networks for biomedical image segmentation. Biomed Opt Express 10(8):3800-3814,2019
Liu P, Zhao H, Li P, Cao F: Automated classification and measurement of fetal ultrasound images with attention feature pyramid network, Second Target Recognition and Artificial Intelligence Summit Forum, SPIE, 2020, p. 114272R
Ni D, Yang Y, Li S, Qin J, Ouyang S, Wang T, Heng PA: Learning based automatic head detection and measurement from fetal ultrasound images via prior knowledge and imaging parameters, 2013 IEEE 10th International Symposium on Biomedical Imaging, IEEE, 2013, pp. 772-775
Zalud I, Good S, Carneiro G, Georgescu B, Aoki K, Green L, Shahrestani F, Okumura R: Fetal biometry: A comparison between experienced sonographers and automated measurements. J Matern Fetal Neonatal Med 22(1):43-50,2009
Satwika IP, Habibie I, Ma'sum MA, Febrian A, Budianto E: Particle swarm optimation based 2-dimensional randomized Hough transform for fetal head biometry detection and approximation in ultrasound imaging, 2014 International Conference on Advanced Computer Science and Information System, IEEE, 2014, pp. 468–473
Ryou H, Yaqub M, Cavallaro A, Papageorghiou AT, Noble JA: Automated 3D ultrasound image analysis for first trimester assessment of fetal health. Phys Med Biol 64(18):185010,2019
Carneiro G, Georgescu B, Good S, Comaniciu D: Detection and measurement of fetal anatomies from ultrasound images using a constrained probabilistic boosting tree. IEEE Trans Med Imaging 27(9):1342-1355,2008
Lu W, Tan J: Detection of incomplete ellipse in images with strong noise by iterative randomized Hough transform (IRHT). Pattern Recognit 41(4):1268-1279,2008
Khanh TLB, Dao DP, Ho NH, Yang HJ, Baek ET, Lee G, Kim SH, Yoo SB: Enhancing U-Net with spatial-channel attention gate for abnormal tissue segmentation in medical imaging. Appl Sci 10(17):5729,2020
Roy AG, Navab N, Wachinger C: Concurrent spatial and channel ‘squeeze & excitation’ in fully convolutional networks, International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI). 2018, 421–429
Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H: Encoder-decoder with atrous separable convolution for semantic image segmentation, Proceedings of the European Conference on Computer Vision (ECCV). 2018, 801–818
Zhao H, Shi J, Qi X, Wang X, Jia J: Pyramid scene parsing network, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2017, 2881–2890
Acknowledgments
The authors would like to thank the anonymous reviewers for their insightful and valuable comments and suggestions.
Funding
This work was supported in part by the National Natural Science Foundation of China (Grant Nos. 11804013, 61871005, 61801312, and 71661167001), the Beijing Natural Science Foundation (Grant No. 4184081), the International Research Cooperation Seed Fund of Beijing University of Technology (Grant No. 2018A15), the Basic Research Fund of Beijing University of Technology, and the Intelligent Physiological Measurement and Clinical Translation, Beijing International Base for Scientific and Technological Cooperation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethical Approval
This retrospective study used fetal ultrasound images provided by the HC18 Challenge, which were collected from the database of the Department of Obstetrics of the Radboud University Medical Center, Nijmegen, the Netherlands, in accordance with the local ethics committee (CMO Arnhem-Nijmegen). All data were anonymized according to the tenets of the Declaration of Helsinki.
Informed Consent
This study used retrospective data provided by the HC18 Challenge, so informed consent was waived.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zeng, Y., Tsui, PH., Wu, W. et al. Fetal Ultrasound Image Segmentation for Automatic Head Circumference Biometry Using Deeply Supervised Attention-Gated V-Net. J Digit Imaging 34, 134–148 (2021). https://doi.org/10.1007/s10278-020-00410-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-020-00410-5