Abstract
Storage and transmission of high-compression 3D radiological images that create high-quality reconstruction upon decompression are critical necessities for effective and efficient teleradiology. To cater to this need, we propose a near lossless 3D image volume compression method based on optimal multilinear singular value decomposition called “3D-VOI-OMLSVD.” The proposed strategy first eliminates any blank 2D image slices from the 3D image volume and uses the selective bounding volume (SBV) to identify and extract the volume of Interest (VOI). Following this, the VOI is decomposed with an optimal multilinear singular value decomposition (OMLSVD) to obtain the corresponding core tensor, factor matrices, and singular values that are compressed with adaptive binary range coder (ABRC), integrated as an entropy encoder. The compressed file can be transferred or transmitted and then decompressed in order to reconstruct the original image. The resultant decompressed VOI is acquired by reversing the above process and then fusing it with the background, using the bound volume coordinates associated with the compressed 3D image. The proposed method performance was tested on a variety of 3D radiological images with different imaging modalities and dimensions using quantitative evaluation metrics such as the compression rate (CR), bit rate (BR), peak signal to noise ratio (PSNR), and structural similarity index (SSIM). Furthermore, we also investigate the impact of VOI extraction on the model performance, before comparing it with two popular compression methods, namely JPEG and JPEG2000. Our proposed method, 3D-VOI-OMLSVD, displayed a high CR value, with a maximum of 37.31, and a low BR, with the lowest reported to be 0.21. The SSIM score was consistently high, with an average performance of 0.9868, while using < 1 second for decoding the image. We observe that with VOI extraction, the compression rate increases manifold, and bit rate drops significantly, and thus reduces the encoding and decoding time to a great extent. Compared to JPEG and JPEG2000, our method consistently performs better in terms of higher CR and lower BR. The results indicate that the proposed compression methodology performs consistently to create high-quality image compressions, and overall gives a better outcome when compared against two state-of-the-art and widely used methods, JPEG and JPEG2000.
Similar content being viewed by others
Introduction
Three dimensional (3D) medical imaging domain has seen great advancement in the past few decades, attributed by the progressively improving medical imaging instrumentation and techniques. There has been a steady increase in the sheer volume of 3D imaging data which is used in clinical radiology for diagnoses or retrospective study and analyses. Hence, organizing, storing, retrieving, and transferring large amounts of imaging data have become a key challenge for healthcare organizations, especially those offering telehealth services. With the growing popularity of telemedicine, further catapulted by the COVID-19 pandemic, the subfield of teleradiology is increasingly becoming popular due to its unique feature of making medical services available, regardless of location and time. However, it comes with its unique set of challenges – expensive technology and service costs, privacy concerns, lack of efficient integration with the electronic health records (EHRs), availability of patient history for continued care outside the network, reimbursement issues, etc. Of all these issues, the one concerned with technology is a critical one. With the discrepancy of limitations of service and equipment available at various healthcare centers across geographical areas of the nation, the issue of sending high-quality medical image is a crucial one, and that’s where efficient medical image compression and decompression plays a very important role in telemedicine.
In the past few decades, there has been an incredible progress in volumetric imaging techniques, especially with dedicated 3D imaging modalities such as computed tomography (CT), magnetic resonance imaging (MRI), positron emitted tomography (PET), and single photon emission computed tomography (SPECT). MRI and CT typically acquire the sequence of 2D cross-sectional images of the organ and constitute the 3D image volume [1] or perform volumetric MRI, whereas PET and SPECT obtain the organ functionality as the third dimension. A wide range of technological innovation has allowed for multimodal imaging of the human anatomy, which also creates a considerable volume of medical image data each year, in all healthcare organizations across the globe [2]. At times, imaging data can be so dense that it easily translates into enormous volumes of data that requires massive storage space along with the need for high-volume transmission, for the purposes of archiving, accessing, and transferring data to ensure accurate and robust diagnostics. These are also the key traits that start defining the captured healthcare data into “big data,” which is classically characterized by the 5 V’s – volume, velocity, variety, value, and veracity. Though there are many floating definitions and features of big data, they are ever evolving as technology advances each year. In the context of imaging data, the “data becomes ‘big’ when image fields are large, pixels are small, frame rates are high, or there are many images acquired per examination” [3]. With the surge of big data volume, with every passing year and the ever-increasing demand for telemedicine, it has become imperative to employ computer-aided diagnosis (CAD) systems to assist radiologists in clinical decision-making [4,5].
Though the emergence of teleradiology has brought about an improvement in the ability to transfer imaging data, there is still a need to improve the supporting data systems to effectively and efficiently handle the voluminous image data collected in the healthcare systems [6]. As a solution towards these complexities, efficient image compression is the ideal strategy to deal with these challenges. In order to transfer and process these data-dense images in large quantities, there is an urgent need to develop an efficient compression technique. There are some notable 3D data compression methods, such as 3D discrete cosine transform (3-D DCT)-based methods [7,8] and 3-D discrete wavelet transforms (3-D DWT) [9,10,11], to name two.
Though impressive compression performance on 3D medical images is desirable, the quality of the decompressed 3D image is also an equally important feature. For this reason, lossless or bit-preserving techniques are the methods of choice for compressing-decompressing medical images for the purpose of transmission [12,13,14]. Since compression performance and quality are inversely related, lossless compression approaches preserve the high-quality images with a low level of compression performance [15, 16]. Due to this unique inverse relation, either the image quality or compression performance must be compromised. Hence, an optimal image compression method which equally improves both factors is preferred [17, 18]. Recent compression models employ deep learning-based techniques [19] which are computationally expensive; see recent review [16]. In this work, we develop an effective and a robust 3D medical image compression technique that incorporates object-based and near lossless properties.
Our approach uses the concept of a tensor decomposition. A tensor is a multidimensional array, with first-order tensor representing a vector and second-order tensor representing a matrix. Generally, an “n”th-order tensor has n basis vectors, and if n ≥ 3, they are referred to as higher-order tensors [20]. Tensor approximation (TA) techniques are widely used to deal with large-scale problems and can be used especially for problems of exponentially growing dimension of data [21]. Tensor-based methods are utilized in various fields, especially in signal and image processing domains, which have adopted tensor-based approaches to a large extent [22,23,24]. Several tensor decomposition types, such as CANDECOMP/PARAFAC (CP) [25,26,27] and Tucker decomposition [28], have been proposed depending on use case. In the signal processing domain, they were initially appended with independent component analysis (ICA) [29] and canonical polyadic decomposition (CPD) [30], both of which are purely tensor-based tools. The decomposition of the tensor may be done with various techniques, but the generalization of singular value decomposition (SVD) for the higher-order tensors is more appropriate. The generalized version of Tucker decomposition is known as multilinear singular value decomposition (MLSVD) [31], and it operates on factor analysis of multidimensional data or model. MLSVD has wide applications due to its efficient high-dimensional computing. The maximum energy packing characteristic of MLSVD leads to promising approximation strategies for image processing applications. Hence, optimized MLSVD is referred to as optimal multilinear singular value decomposition (OMLSVD); the version we will be using in our approach.
In this paper, we propose a compression approach for 3D medical images based on the OMLSVD technique, called the “3D-VOI-OMLSVD.” The initial phase of this technique is a volume of interest (VOI)-based coding where the active volume regions of the original 3D images are identified using a selective bounding volume (SBV) algorithm. The VOI is then subjected to decomposition with OMLSVD, and the compressed data are encoded using the adaptive binary range coder (ABRC) to efficiently encode the compressed information in a near lossless manner. To reconstruct the VOI, the compressed data are then decoded with the ABRC decoder and subjected to inverse OMLSVD. With the aid of SBV coordinate details, the decompressed VOI is then fused with the background to recreate the actual 3D image.
The rest of the paper is structured as follows: Sect. 2 describes the different elements involved in the proposed technique – VOI extraction using SBV algorithm, tensor decomposition using OMLSVD approach, use of ABRC, a binary arithmetic coder technique, for encoding and decoding compressed images, and the dataset used. Section 3 presents the flow of the proposed compression method “3D-VOI-OMLSVD.” Sect. 4 begins with briefly summarizing evaluation metrics used in this study followed by the results from the proposed method, with and without VOI, and later comparing the proposed technique with other popular compression techniques. This section concludes with a brief discussion of future works. Section 5 concludes the paper with a summary of contribution, performance, and limitations.
Materials and Methods
VOI Extraction
Medical images are composed of many elements, and each element has its unique set of characteristics, which can be used for identification. For the purpose of medical image compression, the clinically significant element is known as the region of interest (ROI) in 2D images and as volume of interest (VOI) for 3D images. They are collectively called as object of interest. This object of interest could be brain tumor, traumatic brain injury, blood clot, kidney stone, or any other abnormality in the human body. This region of interest is referred to as the foreground image and is usually captured against the contrast background. In general, most of these 2D and 3D medical images contain a lot of background pixels that carry zero-intensity values. Thus, these background values do not contribute to diagnostics and analyzing process. Separating the foreground pixels from the background pixels can increase the compression performance. The excluded background can be reconstructed during the decompression process to preserve the actual texture and visual quality of the original image. Such object-based coding can improve certain compression algorithms and lead to high fidelity on clinically significant regions.
In this paper, an automated morphological operation-based object detection is proposed, and the VOI is subject to compression. First, the VOI is identified and extracted using the selective bounding volume (SBV) method; it is the 3D extension of the bounding box method. The algorithm of SBV is laid out in detail (algorithm 1). The bounding box operation is applied throughout the volume of 2D images. Corresponding coordinate values of each slice are recorded, and the global values of coordinates are identified for the region of interest in every image slice. Our proposed algorithm ignores any occurring blank image slices that are usually the foremost and endmost slices, because these have no clinically relevant details and are also known as null image slices. Next, the bounding volume is constructed using the acquired global coordinates by applying bounding box morphological operation on the volume, sans the blank images.

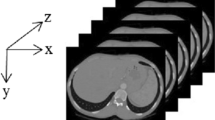
In Fig. 1, the region bounded within the red cube is known as the VOI for the image volume, and the region outside the red cube is the background. This technique excludes any blank 2D slices and includes all slices with the smallest object of interest. This approach allows us to capture the VOIs accurately even in the case of complex 3D images.
Optimized Multilinear Singular Value Decomposition (OMLSVD)
Singular value decomposition (SVD) is one of the tools from numerical analysis that can be used in various image processing tasks including image compression [32, 33]. The concept of SVD is based in linear algebra and has a wide applicability. It is a matrix decomposition/factorization method which uses the eigenvalues to decompose the matrix. It can be mathematically expressed as follows:
here, \(X\) is the original matrix we want to factorize. \(U\) and \(V\) are orthogonal matrices, and the columns of the orthogonal matrix \(U\) are known as left singular vectors (LSVs), and the columns of the orthogonal matrix \(V\) are known as right singular vectors (RSVs). The diagonal matrix \(\Sigma\) contains the eigenvalues in a decreasing order.
SVD is applicable only for 2D data, and the extension of SVD to the higher dimensional data is quite different. Thus, a different approach called the multilinear singular value decomposition (MLSVD) is used for higher dimensional data. MLSVD functions on the basis of tensor multiplication rather than the simple matrix multiplication. Note that the MLSVD was previously known by other names, such as higher-order SVD (HOSVD), N-mode SVD, and Tucker decomposition. The Tucker decomposition was introduced by LR Tucker in 1963 [34]. The basic idea of decomposing a tensor using Tucker decomposition has gone through several modifications and improvements, such as three-mode factor analysis [35], three-mode PCA [36], N-mode PCA [37], higher-order SVD (HOSVD), and N-mode SVD [38]. However, the generalized version is known as the multilinear singular value decomposition (MLSVD) and can be described briefly as follows.
MLSVD decomposes a tensor T \(\in {\mathrm{R}}^{{I}_{1}\times {I}_{2}. . . {I}_{N}}\) into the following components:
-
The core tensor S \(\in {\mathrm{R}}^{{I}_{1}\times {I}_{2}. . . {I}_{N}}\) which is the truncated version of the original tensor with respect to given multilinear rank (\({R}_{1}, {R}_{2}. . . , {R}_{N}\)). Now, the tensor has the dimensions of \({\mathrm{S}}^{{R}_{1}\times {R}_{2} \times . . . \times {R}_{N}}\) where\({R}_{i}\ll {I}_{i } \forall i\).
-
The factor matrices\(U\left\{1\right\}, U\left\{2\right\} . . . . , U\{N\}\), where each factor matrix is unitary and has the dimension \({I}_{n}\times {R}_{n}\) for\(n=1 . . . . . N\).
Thus, we get:
These factor matrices are orthogonal, and the level of interaction between various components can be seen with the entries of core tensor. \({\times }_{n}\) is the mode-n product which indicates the tensor is multiplied in each mode or order. For the illustrative purposes, the 3rd-order tensor with its corresponding decomposition is explained in Fig. 2. Here, a 3rd-order tensor (T) or a volume is decomposed with OMLSVD to get the core tensor S \(\in {\mathrm{R}}^{{I}_{1}\times {I}_{2}\times {I}_{3}}\) with the rank \(({R}_{1} {R}_{2} {R}_{3})\), and the three factor matrices, \(U\left\{1\right\}\in {R}^{{I}_{1}\times {R}_{1}}\), \(U\left\{2\right\}\in {R}^{{I}_{2}\times {R}_{2}}\), and \(U\left\{3\right\}\in {R}^{{I}_{3}\times {R}_{3}}\). The core tensor, S, is multiplied with these factor matrices, \(U\left\{1\right\}, U\left\{2\right\}, and U\{3\}\) using mode-n product to reconstruct the original tensor, T.
Adaptive Binary Range Coder (ABRC)
Typically, frequency domain compression methods need entropy encoding for lossless transmission of the compressed data. The primary idea of data compression is to encode the most frequently occurring symbols with shorter length of codewords and the less frequently occurring symbols to longer length codewords. Hence, arithmetic coding is an efficient entropy coding technique and is used in many transformation-based compression methods [39]. The arithmetic coding process is a symbol-wise recursive algorithm. It encodes and decodes one symbol each iteration. It constructs fractional value codewords which range from 0 to 1. It successively partitions the range to a new interval between the 0 and 1. Hence, the interval is being partitioned on each iteration and comes to a small range of intervals to proceed with. The interval can be specified with the leftmost or rightmost code point and the interval width. The interval width is recorded in order to partition the next interval, and the new code point is computed. With these encoded code strings, we can easily decode the original data in a lossless fashion by reversing the partitions to merge. Our proposed compression method uses the binary arithmetic coder (BAC) to encode the resultant coefficient of the tensor decomposition. BAC is the computational cost effective version of classical arithmetic coders [40]. It is limited to two-element alphabet, of 0 or 1, and it works as a switch-based mechanism. This switch can be defined as a single value. There are several binary arithmetic coders with some modifications, such as M-coder of H.264/H.265 standards [41], MQ-coder from JPEG2000 [42], adaptive BAC based on virtual sliding window [43], adaptive binary range coder (ABRC) [44], and adaptive BAC based on logarithmic domain [45]. Among these coders, ABRC is used in our proposed method because of its efficient encoding capability for multidimensional data. Additionally, it does not require any extra memory for the lookup table. In our method, the coefficients of the core tensor and the magnitudes of the factor matrices are encoded with ABRC to transfer losslessly, which increases the compression performance.
Radiology Imaging Data
The 3D images used for this approach were acquired from the Internet Brain Segmentation Repository (IBSR) [46], an open-source repository by Neuroimaging Tools & Resources Collaboratory (NITRC) in NIfTI (Neuroimaging Informatics Technology Initiative) and DICOM (Digital Imaging and Communications in Medicine) formats. We used twelve 3D image volumes, each of 8-bit depth, of various imaging modalities, to test the performance of 3D-VOI-OMLSVD compression method. Image 1 is a brain MRI (128 × 128 × 27), image 2 is a brain CT (256 × 256 × 99), image 3 is a brain MRI (256 × 256 × 99), images 4 to 11 are skull-stripped brain MRIs (256 × 256 × 63), and image 12 is a chest PET (256 × 256 × 258, obtained from The Cancer Imaging Archive (TCIA)). We identify the core sizes based on the minimum distortion.
Proposed 3D-VOI-OMLSVD Method for Near Lossless Compression
The proposed 3D-VOI-OMLSVD compression method is designed to improve compression performance while maintaining the integrity of image quality. For illustrative purposes, we assume that every 2D medical image fits in the rectangular space for the cross-sectional slice, and the stack of 2D images within the cubic shape has a zero-intensity background around the active regions of interest. The background pixel values are important for storing and transmitting the data, because 8 bits are required for each pixel in case of an image with 8-bit depth. Hence, background is eliminated while extracting the VOI which is the foreground. After eliminating any blank slices, VOI is extracted from the original 3D medical image stack using the SBV VOI detection and extraction technique. The noted global coordinate values of the region of interest for each 2D image slice are used to create the bounding volume. Henceforth, all encoding and decoding processes are applied only to the extracted VOI. After decoding, the whole image is obtained by fusing the VOI to the background using the bounding volume details sent from the encoder.
The extracted VOI is decomposed using MLSVD. VOI is treated as a 3rd-order tensor and gives three factor matrices and the corresponding eigenvalues. Following this, the core tensor is constructed from these eigenvalues with regard to the optimally selected core size. The coefficients of core tensor have a value range of about negative ten thousand to positive ten thousand. Hence, the static assignment of bits for each coefficient is more expensive; i.e., allotting each coefficient with 32 or 64 bits is unnecessary as coefficient values are very small. Thus, we choose to use arithmetic encoding to encode these core tensor coefficients and the factor matrices in order to further reduce the bit rate. The factor matrices U{1}, U{2}, and U{3} are converted as vectors and directly sent to the ABRC. The arithmetic encoded binary sequence binds to the bounding volume coordinates to create a compressed file, which can then be transferred.
In order to reconstruct the original 3D image, the decompression process uses the inverse order of operations that were used for compression. First, arithmetic decoding is performed with the binary sequence to decode the core tensor and the coefficients of the three factor matrices. Then, the VOI is reconstructed by performing tensor product between the core tensor and the factor matrix coefficients. This is a mode-n product. This reconstructs the original VOI, and finally this VOI is fused with the null intensity space of original image volume size to obtain the uncompressed 3D image. The flow of 3D-VOI-OMLSVD compression method is visually depicted in Fig. 3. The illustrations of 3D images are created using 3D Slicer [47, 48].
For example, let us assume a 3D image volume with 256 × 256 × 63 dimensions and 8-bit depth. This means there are 63 cross-sectional 2D image slices that can be collectively seen as a 3D image. As the first step, the VOI is extracted using the SBV method. The dimension size is now reduced to 110 × 113 × 56, as anything outside this VOI is the background null space. The VOI is decomposed with MLSVD to result in a core tensor, eigenvalues, and unitary factor matrices. These values along with the VOI coordinate details and the actual image size are encoded with the arithmetic coding to create the compressed file.
In order to maintain the quality of the reconstructed 3D image, minimum quality distortion is identified by fine-tuning the MLSVD core size. After applying the 3D-VOI-OMLSVD compression method on 3D medical images, we assess the performance using the following quantitative metrics – compression ratio (CR), bit rate (BR), peak signal to noise ratio (PSNR, dB), and structural similarity index (SSIM).
Results and Discussion
Evaluation Metrics
To evaluate the compression performance, we utilize the compression ratio (CR), bit rate (BR)/bits per voxel (BPV), peak signal to noise ratio (PSNR, dB), and structural similarity index (SSIM). Note that these metrics were computed based on the original image bounding box VOI and the compressed image representation of the corresponding VOI, and not the entire image volume.
Compression Ratio (CR)
Compression ratio (CR) is the ratio between the original image size and the compressed image size; i.e., the ratio between number of bits needed to represent the original image and the number of bits needed to represent the compressed image. The CR is computed using the following formula:
where, \({n}_{o}\) – number of bits required to represent the original image.
\({n}_{c}\) – number of bits required to represent the compressed image.
CR values range from 1 to \(\infty\), whereby 1 is the lowest value and \(\infty\) is the highest value. After image compression, higher CR values are desired.
Bit Rate (BR)/Bits per Voxel (BPV)
The average number of bits required per sample of an image is known as the bit rate (BR). It can be computed as the ratio of number of bits needed for representing the compressed image to the total number of unit samples of an image. It can be mathematically represented as follows:
where:
\({n}_{c}\) – number of bits to represent the compressed image.
\({n}_{s}\) – total number of unit samples for an image.
Based on the types of images, the sample unit may vary. For 2D images, the basic unit is a pixel and hence the metric is called bits per pixel (BPP). BPP denotes the number of bits required per pixel of a two-dimensional image where the \({n}_{s}\) denotes the total number of pixels, whereas, in the case of 3D images, the basic unit is known as a voxel, and hence the metric is known as bits per voxel (BPV). BPV represents the number of bits required per voxel in a 3D image where the \({n}_{s}\) denotes the total number of voxels. Both BPP and BPV can be collectively or alternatively called as the bit rate (BR). Compression causes the 8-bit image depth to reduce significantly. Lower BR values are preferred, as it directly correlates to higher compression values.
Peak Signal to Noise Ratio (PSNR)
Peak signal to noise ratio (PSNR, dB) is one of the most widely used evaluation metrics which assess the fidelity of image compression. It is a reference-based evaluation metric that compares the two images in terms of intensity variations in the image. Fundamentally, PSNR (dB) is a ratio between the high peak or maximum intensity value in an image and the intensity changes between the image. The two images for comparison must have the same dimensions. The logarithmic decibel scale is used to express the PSNR values. The mathematical expression for computing PSNR is below:
(or)
where: \({max}_{i}\)– maximum intensity/high peak.
\(MSE\) – mean square error.
In order to calculate the PSNR value, it is essential to calculate the mean square error (\(MSE)\) value. MSE is the intensity difference between the two images, and it uses the Euclidean distance measure. The mathematical expression to compute MSE is:
where: m and n are dimensions of image.
I – original image.
J – decompressed image.
PSNR value can range between 0 to \(\infty\) in decibels (dB). High PSNR value indicates the compared images are highly alike, and the restored image quality is good. If \(MSE\)= 0, the PSNR tends to \(\infty\), which means there is no degradation between the two images.
Structural Similarity Index (SSIM)
Structural similarity index (SSIM) [49] is a human visual system (HVS)-based quality metric which measures the perceptual difference between two images. To compute SSIM, the luminance, contrast, and structural term of an image are multiplied. It is a completely reference-based metric that requires two images which have the same dimensions, namely a reference image and a processed image. This 2D image comparison can be extended to a 3D image too. The SSIM is calculated as below:
where:
\({\mu }_{x}\) and \({\mu }_{y}\) are the local means, \({\sigma }_{x}\) and \({\sigma }_{y}\) are the standard deviations, and \({\sigma }_{xy}\) is cross-covariance for images \(x\) and \(y\). Mean SSIM values range between 0 and 1, and higher values indicate better fidelity between the compared images.
Near Lossless 3D-VOI-OMLSVD Compression Method Performance
We used 12 3D image volumes from the IBSR dataset [45] and TCIA datasets to quantitatively check performance of the technique. Table 1 reports the quantitative evaluation for the above mentioned images, where as Table 2 reports evaluation metrics for the same set of images, with and without extracted VOI.
In Table 1, we observe that the original 3D image is reduced to smaller number of dimensions upon VOI extraction. We were able to obtain the maximum compression ratio (CR) of 37.31:1. The quantitative metrices display promising results, and the high values of SSIM indicate that our approach insures near lossless compression. Conclusively, our approach yields very high accuracy with near lossless compression.
Figure 4 provides a visual comparison between a randomly selected 2D original image slice and the corresponding 2D decompressed image slice, from the twelve 3D image volumes. All the decompressed images display near lossless reconstruction. Image number 3 has the lowest SSIM value of 0.9537 (Table 1) compared to all other images and hence was used as an example image to observe the differences between the original and the decompressed image. In Fig. 5, we show image histograms and corresponding 3D mesh plots of the original, our near lossless 3D-VOI-OMLSVD compression method result, and the difference between them. As can be seen, our method obtains good fidelity to the original uncompressed image.
Representative image (image number 3) was used to observe the visual discrepancy between the original and 3D-VOI-OMLSVD decompressed image. The gray scale (range 0–255) pixel frequency histogram (left) and the corresponding 3D mesh plot (right) are shown in comparison between the original image (top), 3D-VOI-OMLSVD decompressed image (middle), and the difference between the two images (bottom). The mesh plot for difference has been amplified 10 times for better visual comparison
In order to test the effectiveness of the initial step of VOI extraction, we ran our compression method on both, VOI extracted vs non-VOI extracted image stacks. We consistently notice that the optimum core size is lower in VOI extracted approach. The CR values are high, and BR values are significantly low when VOI is extracted. However, we observe similar PSNR and SSIM performance between both approaches.
Preprocessing images using SBV VOI extraction is definitely effective as it reduces the encoding and decoding time for the image, while offering high compression ratio and very similar image reconstruction accuracy.
Comparing with Other Compression Techniques: JPEG, JPEG2000, 3D SPIHT, Huffman Coding, Run Length Coding, Lempel–Ziv-Welch (LZW), and Arithmetic Coding
We compared our proposed compression method with two state-of-the-art international standard (IS) status image compression methods – JPEG [50] and JPEG2000 [51]. JPEG is a standard lossy compression technique that uses RGB channels, whereas JPEG2000 can be used for lossy or lossless compression and can manage up to 256 channels, for images with lower bit rates, such as radiological images; JPEG2000 performs better than JPEG [52]. Although there exists a lossless JPEG LS version which is based on differential pulse-code modulation (DPCM), lossless JPEG2000 is more widely used [53]. In what follows, we detail experimental benchmarking of our 3D-VOI-OMLSVD model against JPEG, JPEG2000, 3D SPIHT, and four popular lossless coding techniques.
We observe an overall higher CR value (Fig. 6) and a lower BR value (Fig. 7) for 3D-VOI-OMLSVD compression method. On average our method requires only 0.43 bit rate or BPV, whereas the JPEG and JPEG2000 need an average of 0.68 and 0.54 BPV, respectively.
We further compared our compression method to the 3D set partitioning in hierarchical trees (3D SPIHT) compression method, using two evaluation metrics, BR/BPV and PSNR (dB). The near lossless 3D-VOI-OMLSVD technique showed overall improved results, across all images, as compared to 3D SPIHT. Our method displayed lower BR/BPV (Fig. 8) and higher PSNR values (Fig. 9). On average, our approach required 0.43 BR/BPV, while 3D SPIHT required 0.46 BR.
Since our technique shows promising performance, almost lossless compression, for all the images (Table 1), we compared it against a few popular ones – Huffman coding, run length coding (RLE), Lempel–Ziv–Welch (LZW), and the arithmetic coding (AC). Compared to all these techniques, our proposed methodology consistently performed the best, with extremely low BR/BPV (Fig. 10).
The time required to retrieve and decode the compressed image is of greater importance than the time required to encode the image. Hence, we also compare the computational run time of our proposed model with four other popular compression methods – Huffman coding, run length coding (RLE), Lempel–Ziv–Welch (LZW), and arithmetic coding (AC) (Table 3). For the entire dataset, we consistently observe that our proposed method takes more time to encode; however, it is the fastest method for decoding, compared to all other tested algorithms. The closest competitor to our 3D-VOI-OMLSVD is the RLE model, which takes the shortest amount of time for encoding and is fairly quick to decode. But our model has a very high compression ratio compared to any other technique.
Future Work
While in this work we compare our proposed technique of near lossless 3D-VOI-OMLSVD compression method with various popular techniques, including two considered as international standards, and very widely used, JPEG and JPEG2000, we would like to further compare it with techniques like 3D DCT and 3D DWT. Since the performance of this approach is promising, we would like to test it on larger datasets and on a variety of other radiological imaging modalities. Using normal and non-normal complex radiological images will also allow us to observe how this 3D-VOI-OMLSVD technique performs when dealing with varying tensor sizes. Though we report the computational time required for encoding and decoding the VOI extracted images, an extension of this work would include the cumulative time frame required for VOI extraction, encoding, decoding, and image reconstruction. Going ahead, we would like to focus on reducing the encoding time using advanced tensor decomposition models.
In this work, we have utilized standard image metrics (PSRN and SSIM) to compare different compression algorithms. To be useful in practice, one needs to use appropriate perceptual evaluation metrics based on human observers [54, 55]. Keeping telehealth in mind, another idea we would like to explore is how constant compression-decompression of the same image affects its quality over time compared to the original image when exchanged between radiology PACS systems. In teleradiology applications, the degradation of image quality over time can be mitigated by transmitting only the compressed version, especially since our proposed model’s decompression operation is very fast.
Conclusion
In this work, we consider a 3D image compression model based on volume of interest (VOI) and optimal multilinear singular value decomposition (3D-VOI-OMLSVD). The proposed 3D-VOI-OMLSVD compression method addresses the need for an effective and efficient 3D medical image compression, as the demand for teleradiology increases. The initial process of VOI extraction improved the performance of our proposed method. We used a tensor-based application of MLSVD to decompose the 3D images in order to obtain the volume of interests (VOIs). Adaptive binary range coder (ABRC) is used for compression, making the image ready for transmittance. In order to reconstruct the original images, this streamlined process is carried out in reverse. We report a low bit rate (BR) and high compression ratio (CR) in comparison to international and widely used standard techniques namely the JPEG and JPEG2000. Our proposed method performs better than 3D set partitioning in hierarchical trees (3D SPIHT) compression method for across different 3D images in the test set. In comparison to a few other state-of-the-art lossless compression methods such as Huffman coding, run length coding, LZW, and arithmetic coding, our method shows significantly low BR, a fairly high encoding time, and the lowest decoding time. The reconstructed images display a very high structural similarity (SSIM) values, indicating we obtain good fidelity to original input 3D data with less loss of information. Our approach with further improvements could find wide applications for all medical image modalities, especially for the purpose of teleradiology and storage [46] where high compression for transmittance and high reconstruction accuracy is required.
References
Peter A. Rinck, Magnetic resonance in medicine: a critical introduction. 2019.
M. Wang, G., Kalra, M., Murugan, V., Xi, Y., Gjesteby, L., Getzin, M., Yang, Q., Cong, W. and Vannier et al., “Vision 20/20: Simultaneous CT‐MRI—Next chapter of multimodality imaging,” Med. Phys., vol. 42, no. 10, pp. 5879–5889, 2015.
M. J. Yaffe, “Emergence of ‘Big Data’ and its potential and current limitations in medical imaging,” Semin. Nucl. Med., vol. 49, no. 2, pp. 94–104, 2019, https://doi.org/10.1053/j.semnuclmed.2018.11.010.
A. A. Abdulla, “Efficient computer-aided diagnosis technique for leukaemia cancer detection,” IET Image Process., vol. 14, no. 17, pp. 4435–4440, 2020.
W. Jorritsma, F. Cnossen, and P. M. van Ooijen, “Improving the radiologist-CAD interaction: designing for appropriate trust,” Clin. Radiol., vol. 70, no. 2, pp. 115–122, 2015.
M. J. McAuliffe, F. M. Lalonde, D. McGarry, W. Gandler, K. Csaky, and B. L. Trus, “Medical image processing, analysis and visualization in clinical research,” in Proceedings 14th IEEE Symposium on Computer-Based Medical Systems. CBMS 2001, 2001, no. February, pp. 381–386, https://doi.org/10.1109/CBMS.2001.941749.
E. Belyaev, “Low bit rate video coding based on three-dimensional discrete pseudo cosine transform,” in International Congress on Ultra Modern Telecommunications and Control Systems, 2010, vol. 1, no. 2, pp. 61–67, https://doi.org/10.1109/ICUMT.2010.5676657.
M. Servais and G. De Jager, “Video compression using the three dimensional discrete cosine transform (3D-DCT),” in Proceedings of the 1997 South African Symposium on Communications and Signal Processing. COMSIG’97, 1997, pp. 27–32, https://doi.org/10.1109/comsig.1997.629976.
B. J. Kim, Z. Xiong, and W. A. Pearlman, “Low bit-rate scalable video coding with 3-D set partitioning in hierarchical trees (3-D SPIHT),” IEEE Trans. Circuits Syst. Video Technol., vol. 10, no. 8, pp. 1374–1387, 2000, https://doi.org/10.1109/76.889025.
E. Belyaev, K. O. Egiazarian, M. Gabbouj, and K. Liu, “A low-complexity joint source-channel videocoding for 3-D DWT codec,” J. Commun., vol. 8, no. 12, pp. 893–901, 2013, https://doi.org/10.12720/jcm.8.12.893-901.
E. Belyaev, K. Egiazarian, and M. Gabbouj, “A low-complexity bit-plane entropy coding and rate control for 3-D DWT based video coding,” IEEE Trans. Multimed., vol. 15, no. 8, pp. 1786–1799, Dec. 2013, https://doi.org/10.1109/TMM.2013.2269315.
P. Kalavathi and S. Boopathiraja, “A medical image compression technique using 2D-DWT with run length encoding,” Glob J Pure Appl Math, vol. 13, no. 5, pp. 87–96, 2017.
S. Boopathiraja and P. Kalavathi, “A near lossless three-dimensional medical image compression technique using 3D-discrete wavelet transform,” Int. J. Biomed. Eng. Technol., vol. 35, no. 3, pp. 191–206, 2021, https://doi.org/10.1504/IJBET.2021.113731.
S. Boopathiraja, P. Kalavathi, and C. Dhanalakshmi, “Significance of image compression and its upshots - A survey,” Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol., vol. 5, no. 2, pp. 1203–1208, Apr. 2019, https://doi.org/10.32628/cseit1952321.
B. Subramanian, K. Palanisamy, and V. B. S. Prasath, “On a hybrid lossless compression technique for three‐dimensional medical images,” J. Appl. Clin. Med. Phys., vol. 22, no. 8, pp. 191–203, Aug. 2021, https://doi.org/10.1002/acm2.12960.
S. Boopathiraja, V. Punitha, P. Kalavathi, and V. B. Prasath, “Computational 2D and 3D medical image data compression models,” Arch. Comput. Methods Eng., pp. 1–33, 2021.
S. S. Parikh, D. Ruiz, H. Kalva, G. Fernández-Escribano, V. Adzic, and Parikh, S.S., Ruiz, D., Kalva, H., Fernández-Escribano, G. and Adzic, V., “High bit-depth medical image compression with hevc,” IEEE J. Biomed. Heal. informatics, vol. 22, no. 2, pp. 552–560, 2017.
S. Abdulla, Alan A and Sellahewa, Harin and Jassim, “Stego quality enhancement by message size reduction and fibonacci bit-plane mapping,” in International conference on research in security standardisation, 2014, pp. 151–166.
S. Ma, X. Zhang, C. Jia, Z. Zhao, S. Wang, and S. Wang, “Image and video compression with neural networks: A review,” IEEE Trans. Circuits Syst. Video Technol., vol. 30, no. 6, pp. 1683–1698, 2020, https://doi.org/10.1109/TCSVT.2019.2910119.
V. De Silva and L. H. Lim, “Tensor rank and the ill-posedness of the best low-rank approximation problem,” SIAM J. Matrix Anal. Appl., vol. 30, no. 3, pp. 1084–1127, 2008, https://doi.org/10.1137/06066518X.
L. Grasedyck, D. Kressner, and C. Tobler, “A literature survey of low-rank tensor approximation techniques,” GAMM Mitteilungen, vol. 36, no. 1, pp. 53–78, Aug. 2013, https://doi.org/10.1002/gamm.201310004.
P. M. Kroonenberg, Applied multiway data analysis. John Wiley & Sons, 2008.
C. Nikias and A. Petropulu, “Higher order spectra analysis, a nonlinear signal processing framework,” Control Eng. Pr., vol. 2, no. 2, pp. 367–368, 1994.
A. Cichocki et al., “Tensor decompositions for signal processing applications: From two-way to multiway component analysis,” IEEE Signal Process. Mag., vol. 32, no. 2, pp. 145–163, 2015.
J. D. Carroll and J.-J. Chang, “Analysis of individual differences in multidimensional scaling via an N-way generalization of ‘Eckart-Young’ decomposition,” Psychometrika, vol. 35, no. 3, pp. 283–319, 1970, https://doi.org/10.1007/BF02310791.
R. Harshman, “Foundations of the PARAFAC procedure: Models and conditions for an ‘explanatory’ multimodal factor analysis,” UCLA Work. Pap. Phonetics, vol. 16, no. 10, pp. 1–84, 1970, [Online]. Available: http://www.psychology.uwo.ca/faculty/harshman/wpppfac0.pdf.
T. G. Kolda and B. W. Bader, “Tensor decompositions and applications,” SIAM Rev., vol. 51, no. 3, pp. 455–500, 2009.
V. Bhatt, S. Kumar, and S. Saini, “Tucker decomposition and applications,” Mater. Today Proc., vol. 46, pp. 10787–10792, 2021.
P. Comon, “Independent component analysis, a new concept?,” Signal Processing, vol. 36, no. 3, pp. 287–314, 1994.
N. D. Sidiropoulos, R. Bro, and G. B. Giannakis, “Parallel factor analysis in sensor array processing,” IEEE Trans. Signal Process., vol. 48, no. 8, pp. 2377–2388, 2000.
L. De Lathauwer, B. De Moor, J. Vandewalle, L. de Lathauwer, B. De Moor, and J. Vandewalle, “A multilinear singular value decomposition,” SIAM J. Matrix Anal. Appl., vol. 21, no. 4, pp. 1253–1278, 2000.
H. C. Andrews and C. L. Patterson, “Singular value decompositions and digital image processing,” IEEE Trans. Acoust., vol. 24, no. 1, pp. 26–53, 1976, https://doi.org/10.1109/TASSP.1976.1162766.
J.-F. Yang and C.-L. Lu, “Combined techniques of singular value decomposition and vector quantization for image coding,” IEEE Trans. image Process., vol. 4, no. 8, pp. 1141–1146, 1995, https://doi.org/10.1109/83.403419.
L. R. Tucker, “Implications of factor analysis of three-way matrices for measurement of change,” Probl. Meas. Chang., vol. 15, no. 122–137, p. 3, 1963.
L. R. Tucker, “Some mathematical notes on three-mode factor analysis,” Psychometrika, vol. 31, no. 3, pp. 279–311, 1966.
P. M. Kroonenberg and J. De Leeuw, “Principal component analysis of three-mode data by means of alternating least squares algorithms,” Psychometrika, vol. 45, no. 1, pp. 69–97, 1980.
A. Kapteyn, H. Neudecker, and T. Wansbeek, “An approach ton-mode components analysis,” Psychometrika, vol. 51, no. 2, pp. 269–275, Jun. 1986, https://doi.org/10.1007/BF02293984.
O. Vasilescu, M. Alex, and D. Terzopoulos, “Multilinear analysis of image ensembles: Tensorfaces,” in European conference on computer vision, 2002, vol. 2350, pp. 447–460.
I. Sebestyen, “JPEG: Still image data compression standard,” Comput. Stand. Interfaces, vol. 15, no. 4, pp. 365–366, Sep. 1993, https://doi.org/10.1016/0920-5489(93)90038-S.
G. Langdon, “An introduction to arithmetic coding,” IBM J. Res. Dev., vol. 28, no. 2, pp. 135–149, 1984, https://doi.org/10.1147/rd.282.0135.
R. Osorio and J. D. Bruguera, “High-throughput architecture for H. 264/AVC CABAC compression system,” IEEE Trans. Circuits Syst. Video Technol., vol. 16, no. 11, pp. 1376–1384, 2006.
ITU-T and ISO/IEC JTC 1, “JPEG 2000 image coding system: core coding cystem, ITU-T recommendation T.800 and ISO/IEC 15444–1,” 2000.
E. Belyaev, A. Turlikov, K. Egiazarian, and M. Gabbouj, “An efficient adaptive binary arithmetic coder with low memory requirement,” IEEE J. Sel. Top. Signal Process., vol. 7, no. 6, pp. 1053–1061, 2013.
E. Belyaev, Kai Liu, M. Gabbouj, and YunSong Li, “An efficient adaptive binary range coder and its VLSI architecture,” IEEE Trans. Circuits Syst. Video Technol., vol. 25, no. 8, pp. 1435–1446, Aug. 2015, https://doi.org/10.1109/TCSVT.2014.2372291.
Q. Yu, W. Yu, P. Yang, J. Zheng, X. Zheng, and Y. He, “An efficient adaptive binary arithmetic coder based on logarithmic domain,” IEEE Trans. image Process., vol. 24, no. 11, pp. 4225–4239, 2015.
“IBSR.” https://www.nitrc.org/projects/ibsr/ (Accessed 11 May 2021).
R. Kikinis, S. D. Pieper, and K. G. Vosburgh, “3D Slicer: a platform for subject-specific image analysis, visualization, and clinical support,” in Intraoperative imaging and image-guided therapy, Springer, 2014, pp. 277–289.
“3D Slicer.” https://www.slicer.org/ (Accessed 11 May 2021).
Z. Wang, A. C. Bovik, H. R. Sheikh, and E. P. Simoncelli, “Image quality assessment: from error visibility to structural similarity,” IEEE Trans. image Process., vol. 13, no. 4, pp. 600–612, 2004.
G. K. Wallace, “The JPEG still picture compression standard,” IEEE Trans. Consum. Electron., vol. 38, no. 1, pp. xviii–xxxiv, 1992.
A. Skodras, C. Christopoulos, and T. Ebrahimi, “The jpeg 2000 still image compression standard,” IEEE Signal Process. Mag., vol. 18, no. 5, pp. 36–58, 2001.
S. Grgic, M. Mrak, and M. Grgic, “Comparison of JPEG image coders,” Univ. Zagreb. Fac. Electr. Eng. Comput. Unska, vol. 3, 2001.
G. Hudson, A. Leger, B. Niss, and I. Sebestyen, “JPEG at 25: Still Going Strong,” IEEE Multimed., vol. 24, no. 2, pp. 96–103, 2017, https://doi.org/10.1109/MMUL.2017.38.
M. L. Mele, D. Millar, and C. E. Rijnders, “Validating a quality perception model for image compression: The subjective evaluation of the cogisen’s image compression plug-in,” Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 9731, pp. 350–359, 2016, https://doi.org/10.1007/978-3-319-39510-4_33.
Y. Patel, S. Appalaraju, and R. Manmatha, “Human Perceptual Evaluations for Image Compression,” pp. 1–5, 2019, [Online]. Available: http://arxiv.org/abs/1908.04187.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Boopathiraja, S., Kalavathi, P., Deoghare, S. et al. Near Lossless Compression for 3D Radiological Images Using Optimal Multilinear Singular Value Decomposition (3D-VOI-OMLSVD). J Digit Imaging 36, 259–275 (2023). https://doi.org/10.1007/s10278-022-00687-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-022-00687-8