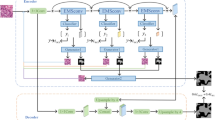
Abstract
The class distribution of a training dataset is an important factor which influences the performance of a deep learning-based system. Understanding the optimal class distribution is therefore crucial when building a new training set which may be costly to annotate. This is the case for histological images used in cancer diagnosis where image annotation requires domain experts. In this paper, we tackle the problem of finding the optimal class distribution of a training set to be able to train an optimal model that detects cancer in histological images. We formulate several hypotheses which are then tested in scores of experiments with hundreds of trials. The experiments have been designed to account for both segmentation and classification frameworks with various class distributions in the training set, such as natural, balanced, over-represented cancer, and over-represented non-cancer. In the case of cancer detection, the experiments show several important results: (a) the natural class distribution produces more accurate results than the artificially generated balanced distribution; (b) the over-representation of non-cancer/negative classes (healthy tissue and/or background classes) compared to cancer/positive classes reduces the number of samples which are falsely predicted as cancer (false positive); (c) the least expensive to annotate non-ROI (non-region-of-interest) data can be useful in compensating for the performance loss in the system due to a shortage of expensive to annotate ROI data; (d) the multi-label examples are more useful than the single-label ones to train a segmentation model; and (e) when the classification model is tuned with a balanced validation set, it is less affected than the segmentation model by the class distribution of the training set.
















Similar content being viewed by others
Notes
A histological slide is a microscopic examination of tissue used by physicians to study the manifestations of disease.
Natural distribution is the distribution a data originally has, which can be either balanced or biased to a certain class.
Sensitivity is the proportion of actual positive cases that are predicted as positive.
Negative example wrongly predicted as positive class
References
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: F. Pereira, C.J.C. Burges, L. Bottou, K.Q. Weinberger (eds.) Advances in Neural Information Processing Systems 25, pp. 1097–1105. Curran Associates, Inc. (2012)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521(7553), 436–444 (2015). 10.1038/nature14539. URL http://dx.doi.org/10.1038/nature14539
Rawat, W., Wang, Z.: Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput. 29(9), 2352–2449 (2017). 10.1162/neco-a-00990. URL https://doi.org/10.1162/neco-a-00990
Bejnordi, B.E., Veta, M., Van Diest, P.J., Van Ginneken, B., Karssemeijer, N., Litjens, G., Van Der Laak, J.A., Hermsen, M., Manson, Q.F., Balkenhol, M., et al.: Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 318(22), 2199–2210 (2017)
Liu, Y., Gadepalli, K.K., Norouzi, M., Dahl, G., Kohlberger, T., Venugopalan, S., Boyko, A.S., Timofeev, A., Nelson, P.Q., Corrado, G., Hipp, J., Peng, L., Stumpe, M.: Detecting cancer metastases on gigapixel pathology images (2017). URL https://arxiv.org/abs/1703.02442. Initial publication on arxiv, then submit to MICCAI
Wang, D., Khosla, A., Gargeya, R., Irshad, H., Beck, A.H.: Deep learning for identifying metastatic breast cancer. CoRR abs/1606.05718 (2016). URL http://dblp.uni-trier.de/db/journals/corr/corr1606.html#WangKGIB16
Gurcan, M.N., Boucheron, L.E., Can, A., Madabhushi, A., Rajpoot, N.M., Yener, B.: Histopathological image analysis: A review. IEEE reviews in biomedical engineering 2, 147–171 (2009)
Hu, Z., Tang, J., Wang, Z., Zhang, K., Zhang, L., Sun, Q.: Deep learning for image-based cancer detection and diagnosis- a survey. Pattern Recognition 83, 134–149 (2018)
Komura, D., Ishikawa, S.: Machine learning methods for histopathological image analysis. Computational and Structural Biotechnology Journal 16, 34–42 (2018)
Litjens, G., Kooi, T., Bejnordi, B.E., Setio, A.A.A., Ciompi, F., Ghafoorian, M., Van Der Laak, J.A., Van Ginneken, B., Sánchez, C.I.: A survey on deep learning in medical image analysis. Medical image analysis 42, 60–88 (2017)
Hinz, T., Navarro-Guerrero, N., Magg, S., Wermter, S.: Speeding up the hyperparameter optimization of deep convolutional neural networks. International Journal of Computational Intelligence and Applications 17(02), 1850008 (2018)
Cracknell, M.J., Reading, A.M.: Geological mapping using remote sensing data: A comparison of five machine learning algorithms, their response to variations in the spatial distribution of training data and the use of explicit spatial information. Computers & Geosciences 63, 22–33 (2014)
Crawford, K.: Artificial intelligence’s white guy problem. The New York Times 25(06) (2016)
Batista, G.E., Prati, R.C., Monard, M.C.: A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD explorations newsletter 6(1), 20–29 (2004)
Buda, M., Maki, A., Mazurowski, M.A.: A systematic study of the class imbalance problem in convolutional neural networks. Neural Networks 106, 249–259 (2018)
Prati, R.C., Batista, G.E., Silva, D.F.: Class imbalance revisited: a new experimental setup to assess the performance of treatment methods. Knowledge and Information Systems 45(1), 247–270 (2015)
Weiss, G.M., Provost, F.: Learning when training data are costly: The effect of class distribution on tree induction. Journal of Artificial Intelligence Research 19, 315–354 (2003)
Chawla, N.V., Bowyer, K.W., Hall, L.O., Kegelmeyer, W.P.: Smote: synthetic minority over-sampling technique. Journal of artificial intelligence research 16, 321–357 (2002)
Khan, S.H., Hayat, M., Bennamoun, M., Sohel, F.A., Togneri, R.: Cost-sensitive learning of deep feature representations from imbalanced data. IEEE transactions on neural networks and learning systems (2017)
Halicek, M., Shahedi, M., Little, J.V., Chen, A.Y., Myers, L.L., Sumer, B.D., Fei, B.: Head and neck cancer detection in digitized whole-slide histology using convolutional neural networks. Scientific reports 9(1), 1–11 (2019)
Weiss, G.M., Provost, F.: The effect of class distribution on classifier learning: an empirical study. Rutgers Univ (2001)
Zhu, Z., Gallant, A.L., Woodcock, C.E., Pengra, B., Olofsson, P., Loveland, T.R., Jin, S., Dahal, D., Yang, L., Auch, R.F.: Optimizing selection of training and auxiliary data for operational land cover classification for the lcmap initiative. ISPRS Journal of Photogrammetry and Remote Sensing 122, 206–221 (2016)
Pham, H.H.N., Futakuchi, M., Bychkov, A., Furukawa, T., Kuroda, K., Fukuoka, J.: Detection of lung cancer lymph node metastases from whole-slide histopathologic images using a two-step deep learning approach. The American journal of pathology 189(12), 2428–2439 (2019)
Everingham, M., Gool, L., Williams, C.K., Winn, J., Zisserman, A.: The pascal visual object classes (voc) challenge. Int. J. Comput. Vision 88(2), 303–338 (2010). 10.1007/s11263-009-0275-4. URL http://dx.doi.org/10.1007/s11263-009-0275-4
Noh, H., Hong, S., Han, B.: Learning deconvolution network for semantic segmentation. In: Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), ICCV ’15, pp. 1520–1528. IEEE Computer Society, Washington, DC, USA (2015). 10.1109/ICCV.2015.178. URL http://dx.doi.org/10.1109/ICCV.2015.178
Afzal, S., Maqsood, M., Nazir, F., Khan, U., Aadil, F., Awan, K.M., Mehmood, I., Song, O.Y.: A data augmentation-based framework to handle class imbalance problem for alzheimer’s stage detection. IEEE Access 7, 115528–115539 (2019)
Chawla, N.V., Lazarevic, A., Hall, L.O., Bowyer, K.W.: Smoteboost: Improving prediction of the minority class in boosting. In: European conference on principles of data mining and knowledge discovery, pp. 107–119. Springer (2003)
Jaccard, N., Rogers, T.W., Morton, E.J., Griffin, L.D.: Detection of concealed cars in complex cargo x-ray imagery using deep learning. Journal of X-ray Science and Technology 25(3), 323–339 (2017)
Kubat, M., Matwin, S., et al.: Addressing the curse of imbalanced training sets: one-sided selection. In: Icml, vol. 97, pp. 179–186. Nashville, USA (1997)
Levi, G., Hassner, T.: Age and gender classification using convolutional neural networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 34–42 (2015)
Masko, D., Hensman, P.: The impact of imbalanced training data for convolutional neural networks (2015)
Sun, Y., Kamel, M.S., Wong, A.K., Wang, Y.: Cost-sensitive boosting for classification of imbalanced data. Pattern Recognition 40(12), 3358–3378 (2007)
Wang, S., Liu, W., Wu, J., Cao, L., Meng, Q., Kennedy, P.J.: Training deep neural networks on imbalanced data sets. In: Neural Networks (IJCNN), 2016 International Joint Conference on, pp. 4368–4374. IEEE (2016)
Wu, Y., Ding, Y., Feng, J.: Smote-boost-based sparse bayesian model for flood prediction. EURASIP Journal on Wireless Communications and Networking 2020, 1–12 (2020)
Yuan, X., Xie, L., Abouelenien, M.: A regularized ensemble framework of deep learning for cancer detection from multi-class, imbalanced training data. Pattern Recognition 77, 160–172 (2018)
Johnson, J.M., Khoshgoftaar, T.M.: Survey on deep learning with class imbalance. Journal of Big Data 6(1), 27 (2019)
Hamad, R.A., Kimura, M., Lundström, J.: Efficacy of imbalanced data handling methods on deep learning for smart homes environments. SN Computer Science 1(4), 1–10 (2020)
Baloch, B.K., Kumar, S., Haresh, S., Rehman, A., Syed, T.: Focused anchors loss: Cost-sensitive learning of discriminative features for imbalanced classification. In: Asian Conference on Machine Learning, pp. 822–835 (2019)
Havaei, M., Davy, A., Warde-Farley, D., Biard, A., Courville, A., Bengio, Y., Pal, C., Jodoin, P.M., Larochelle, H.: Brain tumor segmentation with deep neural networks. Medical image analysis 35, 18–31 (2017)
Rendón, E., Alejo, R., Castorena, C., Isidro-Ortega, F.J., Granda-Gutiérrez, E.E.: Data sampling methods to deal with the big data multi-class imbalance problem. Applied Sciences 10(4), 1276 (2020)
Reshma, I.A., Cussat-Blanc, S., Ionescu, R.T., Luga, H., Mothe, J.: Natural vs balanced distribution in deep learning on whole slide images for cancer detection. In: Proceedings of the 36th Annual ACM Symposium on Applied Computing, pp. 18–25 (2021)
Dheeru, D., Karra Taniskidou, E.: UCI machine learning repository (2017). URL http://archive.ics.uci.edu/ml
Wolpert, D.H.: The lack of a priori distinctions between learning algorithms. Neural Computation 8(7), 1341–1390 (1996)
Liu, Y., Kohlberger, T., Norouzi, M., Dahl, G.E., Smith, J.L., Mohtashamian, A., Olson, N., Peng, L.H., Hipp, J.D., Stumpe, M.C.: Artificial intelligence–based breast cancer nodal metastasis detection: Insights into the black box for pathologists. Archives of pathology & lab. medicine (2018)
Bera, K., Schalper, K.A., Rimm, D.L., Velcheti, V., Madabhushi, A.: Artificial intelligence in digital pathology new tools for diagnosis and precision oncology. Nature reviews Clinical oncology 16(11), 703–715 (2019)
Fan, K., Wen, S., Deng, Z.: Deep learning for detecting breast cancer metastases on wsi. In: Innovation in Medicine and Healthcare Systems, and Multimedia, pp. 137–145. Springer (2019)
Lin, H., Chen, H., Dou, Q., Wang, L., Qin, J., Heng, P.A.: Scannet: A fast and dense scanning framework for metastastic breast cancer detection from whole-slide image. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), pp. 539–546. IEEE (2018)
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V., Rabinovich, A.: Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015)
Veeling, B.S., Linmans, J., Winkens, J., Cohen, T., Welling, M.: Rotation equivariant cnns for digital pathology. In: International Conference on Medical image computing and computer-assisted intervention, pp. 210–218. Springer (2018)
Cohen, T., Welling, M.: Group equivariant convolutional networks. In: International conference on machine learning, pp. 2990–2999 (2016)
Mejbri, S.: Deep learning applied to multivariate medical data. PhD dissertation, Universite Toulouse III-Paul Sabatier (2019)
Mejbri, S., Franchet, C., Reshma, I.A., Mothe, J., Brousset, P., Faure, E.: Deep analysis of cnn settings for new cancer whole-slide histological images segmentation: the case of small training sets. In: 6th International Conference on Bioimaging (2019)
Zhou, X., Li, C., Rahaman, M.M., Yao, Y., Ai, S., Sun, C., Wang, Q., Zhang, Y., Li, M., Li, X., et al.: A comprehensive review for breast histopathology image analysis using classical and deep neural networks. IEEE Access 8, 90931–90956 (2020)
Farahani, N.: Whole slide imaging in pathology: advantages, limitations, and emerging perspectives (2015)
Kumar, N., Gupta, R., Gupta, S.: Whole slide imaging (wsi) in pathology: Current perspectives and future directions. Journal of Digital Imaging (2020)
Alexi, B., Altuna, H., Babak, B.E., Wauters Carla, Geert, L., Jeroen, L.V., Dijk Van Marcory, Maschenka, B., Meyke, H., Nikolas, S., Oscar, G., Paul, D.V., Peter, B., Bult Peter, Manson Quirine, Vogels Rob, Rob, D.L.V.: Supporting data for 1399 h&e-stained sentinel lymph node sections of breast cancer patients: the camelyon dataset (2018). 10.5524/100439. URL http://gigadb.org/dataset/100439
Walach, E., Wolf, L.: Learning to count with cnn boosting. In: European Conference on Computer Vision, pp. 660–676. Springer (2016)
Shorten, C., Khoshgoftaar, T.M.: A survey on image data augmentation for deep learning. Journal of Big Data 6(1), 60 (2019)
Kellenberger, B., Marcos, D., Tuia, D.: Detecting mammals in uav images: Best practices to address a substantially imbalanced dataset with deep learning. Remote sensing of environment 216, 139–153 (2018)
Koller, O., Ney, H., Bowden, R.: Deep learning of mouth shapes for sign language. In: Proceedings of the IEEE International Conference on Computer Vision Workshops, pp. 85–91 (2015)
Lee, D.K., In, J., Lee, S.: Standard deviation and standard error of the mean. Korean journal of anesthesiology 68(3), 220 (2015)
Ronneberger, O., P.Fischer, Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI), LNCS, 9351, 234–241. Springer (2015). URL http://lmb.informatik.uni-freiburg.de/Publications/2015/RFB15a
Chollet, F., et al.: Keras. https://keras.io (2015)
Kingma, D.P., Ba, J.: Adam: A method for stochastic optimization. In: Proceedings of ICLR (2015)
Graham, S., Epstein, D., Rajpoot, N.: Dense steerable filter cnns for exploiting rotational symmetry in histology images. IEEE Transactions on Medical Imaging (2020)
Bylinskii, Z., Judd, T., Oliva, A., Torralba, A., Durand, F.: What do different evaluation metrics tell us about saliency models? IEEE transactions on pattern analysis and machine intelligence 41(3), 740–757 (2018)
Davis, J., Goadrich, M.: The relationship between precision-recall and roc curves. In: Proceedings of the 23rd international conference on Machine learning, pp. 233–240 (2006)
Acknowledgements
The authors would like to thank Dr. Md Zia Ullah for his fruitful discussions. Part of this research has received funding from the NO Grants 2014-2021, under project ELO-Hyp contract no. 24/2020.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Reshma, I.A., Franchet, C., Gaspard, M. et al. Finding a Suitable Class Distribution for Building Histological Images Datasets Used in Deep Model Training—The Case of Cancer Detection. J Digit Imaging 35, 1326–1349 (2022). https://doi.org/10.1007/s10278-022-00618-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-022-00618-7