Abstract
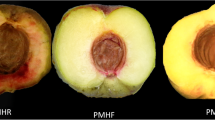
‘Red Meat Honey Crisp (RMHC)’ has been widely cultivated by growers in recent years due to its early maturity, and red meat type characteristics. As a bud variant of ‘Super Red (SR)’ peach, red flesh is the most distinctive characteristic of ‘Red Meat Honey Crisp (RMHC)’. However, the mechanism of red flesh formation in ‘RMHC’ remains unclear. In this study, 79 differentially produced metabolites were identified by metabolomics analysis. The anthocyanin content in ‘RMHC’ was significantly higher than that in ‘SR’ during the same period, such as cyanidin O-syringic acid and cyanidin 3-O-glucoside. Other flavonoids also increased during the formation of red flesh, including flavonols (6-hydroxykaempferol-7-O-glucoside, hyperin), flavanols (protocatechuic acid, (+)-gallocatechin), and flavonoids (chrysoeriol 5-O-hexoside, tricetin). In addition, transcriptomic analysis and RT-qPCR showed that the expression levels of the flavonoid synthesis pathway transcription factor MYB75 and some structural genes, such as PpDFR, PpCHS, PpC4H, and PpLDOX increased significantly in ‘RMHC’. Subcellular localization analysis revealed that MYB75 was localized to the nucleus. Yeast single hybridization assays showed that MYB75 bound to the cis-acting element CCGTTG of the PpDFR promoter region. The MYB75-PpDFR regulatory network was identified to be a key pathway in the reddening of ‘RMHC’ flesh. Moreover, this is the first study to describe the cause for red meat reddening in ‘RMHC’ compared to ‘SR’ peaches using transcriptomics, metabolomics and molecular methods. Our study identified a key transcription factor involved in the regulation of the flavonoid synthetic pathway and contributes to peach breeding-related efforts as well as the identification of genes involved in color formation in other species.







Similar content being viewed by others
Data Availability
Data are available on request to the corresponding author. The transcriptome sequencing has been deposited under BioProjectin NCBI. The accession number for these SRA data is PRJNA1038688.
References
Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26:962–980. https://doi.org/10.1105/tpc.113.122069
An JP, Wang XF, Zhang XW, Xu HF, Bi SQ, You CX, Hao YJ (2020) An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J 18:337–353. https://doi.org/10.1111/pbi.13201
Araguirang GE, Richter AS (2022) Activation of anthocyanin biosynthesis in high light - what is the initial signal? New Phytol 236:2037–2043. https://doi.org/10.1111/nph.18488
Broucke E, Dang TTV, Li Y, Hulsmans S, Van Leene J, De Jaeger G, Hwang I, Wim V, Rolland F (2023) SnRK1 inhibits anthocyanin biosynthesis through both transcriptional regulation and direct phosphorylation and dissociation of the MYB/bHLH/TTG1 MBW complex. Plant J. https://doi.org/10.1111/tpj.16312
Cao K, Ding T, Mao D, Zhu G, Fang W, Chen C, Wang X, Wang L (2018) Transcriptome analysis reveals novel genes involved in anthocyanin biosynthesis in the flesh of peach. Plant Physiol Biochem 123:94–102. https://doi.org/10.1016/j.plaphy.2017.12.005
Cao Y, Xie L, Ma Y, Ren C, Xing M, Fu Z, Wu X, Yin X, Xu C, Li X (2019) PpMYB15 and PpMYBF1 transcription factors are involved in regulating flavonol biosynthesis in peach fruit. J Agric Food Chem 67:644–652. https://doi.org/10.1021/acs.jafc.8b04810
Chagné D, Lin-Wang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, Micheletti D, McGhie T, Crowhurst RN, Storey RD, Velasco R, Hellens RP, Gardiner SE, Allan AC (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161:225–239. https://doi.org/10.1104/pp.112.206771
Dare AP, Schaffer RJ, Lin-Wang K, Allan AC, Hellens RP (2008) Identification of a cis-regulatory element by transient analysis of co-ordinately regulated genes. Plant Methods 4:17. https://doi.org/10.1186/1746-4811-4-17
Drogoudi P, Gerasopoulos D, Kafkaletou M, Tsantili E (2017) Phenotypic characterization of qualitative parameters and antioxidant contents in peach and nectarine fruit and changes after jam preparation. J Sci Food Agric 97:3374–3383. https://doi.org/10.1002/jsfa.8188
Es-Safi NE, Cheynier V, Moutounet M (2002) Interactions between cyanidin 3-O-glucoside and furfural derivatives and their impact on food color changes. J Agric Food Chem 50:5586–5595. https://doi.org/10.1021/jf025504q
Falchi R, Vendramin E, Zanon L, Scalabrin S, Cipriani G, Verde I, Vizzotto G, Morgante M (2013) Three distinct mutational mechanisms acting on a single gene underpin the origin of yellow flesh in peach. Plant J 76:175–187. https://doi.org/10.1111/tpj.12283
Feller A, Yuan L, Grotewold E (2017) The BIF Domain in plant bHLH proteins is an ACT-like domain. Plant Cell 29:1800–1802. https://doi.org/10.1105/tpc.17.00356
Feng SQ, Wang YL, Yang S, Xu YT, Chen XS (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232:245–255. https://doi.org/10.1007/s00425-010-1170-5
Feng RC, Zhang CH, Ma RJ, Cai ZX, Lin Y, Yu ML (2019) Identification and characterization of WD40 superfamily genes in peach. Gene 710:291–306. https://doi.org/10.1016/j.gene.2019.06.010
Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT (2015) Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol 33:243–246. https://doi.org/10.1038/nbt.3172
Hu C, Gong Y, Jin S, Zhu Q (2011) Molecular analysis of a UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT) gene from purple potato (Solanum tuberosum). Mol Biol Rep 38:561–567. https://doi.org/10.1007/s11033-010-0141-z
Hui Z, Kui LW, Wang FR, Espley RV, Ren F, Zhao JB, Ogutu C, He H, Jiang Q, Allan AC, Han YP (2019) Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol 221:1919–1934. https://doi.org/10.1111/nph.15486
Ji XL, Ren J, Zhang YX, Lang SY, Wang D, Song XS (2021) Integrated analysis of the metabolome and transcriptome on anthocyanin biosynthesis in four developmental stages of Cerasus Humilis peel coloration. Int J Mol Sci 22. https://doi.org/10.3390/ijms222111880
Jiang C, Wen Q, Chen Y, Xu LA, Huang MR (2011) Efficient extraction of RNA from various Camellia species rich in secondary metabolites for deep transcriptome sequencing and gene expression analysis. Afr J Biotechnol 10:16769–16773. https://doi.org/10.5897/ajb11.235
Karak P (2019) Biological activities of flavonoids: an overview. Int J Pharm Sci Res 10:1567–1574. https://doi.org/10.13040/ijpsr.0975-8232.10
Khan IA, Cao K, Guo J, Li Y, Wang Q, Yang X, Wu J, Fang W, Wang L (2022a) Identification of key gene networks controlling anthocyanin biosynthesis in peach flower. Plant Sci 316:111151. https://doi.org/10.1016/j.plantsci.2021.111151
Khan IA, Rahman MU, Sakhi S, Nawaz G, Khan AA, Ahmad T, Adnan M, Khan SM (2022b) PpMYB39 activates PpDFR to modulate anthocyanin biosynthesis during peach fruit maturation. Horticulturae 8. https://doi.org/10.3390/horticulturae8040332
Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N (2001) Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci 160:543–550. https://doi.org/10.1016/s0168-9452(00)00425-8
Kreynes AE, Yong ZH, Ellis BE (2021) Developmental phenotypes of Arabidopsis plants expressing phosphovariants of AtMYB75. Plant Signal Behav 16. https://doi.org/10.1080/15592324.2020.1836454
LaFountain AM, Yuan YW (2021) Repressors of anthocyanin biosynthesis. New Phytol 231:933–949. https://doi.org/10.1111/nph.17397
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50. https://doi.org/10.1186/1471-2229-10-50
Liu XF, Feng C, Zhang MM, Yin XR, Xu CJ, Chen KS (2013) The MrWD40-1 gene of Chinese bayberry (Myrica rubra) interacts with MYB and bHLH to enhance anthocyanin accumulation. Plant Mol Biol Rep 31:1474–1484. https://doi.org/10.1007/s11105-013-0621-0
Liu Y, Tikunov Y, Schouten RE, Marcelis LFM, Visser RGF, Bovy A (2018) Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: a review. Front Chem 6:52. https://doi.org/10.3389/fchem.2018.00052
Liu H, Xiong JS, Jiang YT, Wang L, Cheng ZM (2019a) Evolution of the R2R3-MYB gene family in six Rosaceae species and expression in woodland strawberry. J Integr Agric 18:2753–2770. https://doi.org/10.1016/s2095-3119(19)62818-2
Liu X, Chen M, Wen BB, Fu XL, Li DM, Chen XD, Gao DS, Li L, Xiao W (2019b) Transcriptome analysis of peach (Prunus persica) fruit skin and differential expression of related pigment genes. Sci Hortic 250:271–277. https://doi.org/10.1016/j.scienta.2019.02.058
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Millar DJ, Long M, Donovan G, Fraser PD, Boudet AM, Danoun S, Bramley PM, Bolwell GP (2007) Introduction of sense constructs of cinnamate 4-hydroxylase (CYP73A24) in transgenic tomato plants shows opposite effects on flux into stem lignin and fruit flavonoids. Phytochemistry 68:1497–1509. https://doi.org/10.1016/j.phytochem.2007.03.018
Montefiori M, Espley RV, Stevenson DD, Cooney JM, Datson PM, Saiz A, Atkinson RG, McGhie T, Hellens RP, Allan AC (2011) The control of kiwifruit red flesh colour. In: 7th International Symposium on Kiwifruit, Faenza, ITALY, Nov 25 2011. Acta Horticulturae. pp 103–109. https://doi.org/10.17660/ActaHortic.2011.913.11
Rahim MA, Busatto N, Trainotti L (2014) Regulation of anthocyanin biosynthesis in peach fruits. Planta 240:913–929. https://doi.org/10.1007/s00425-014-2078-2
Seo MS, Kim JS (2017) Understanding of MYB transcription factors involved in glucosinolate biosynthesis in Brassicaceae. Molecules 22. https://doi.org/10.3390/molecules22091549
Springob K, Nakajima J, Yamazaki M, Saito K (2003) Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 20:288–303. https://doi.org/10.1039/b109542k
Sun RZ, Cheng G, Li Q, Zhu YR, Zhang X, Wang Y, He YN, Li SY, He L, Chen W, Pan QH, Duan CQ, Wang J (2019) Comparative physiological, metabolomic, and transcriptomic analyses reveal developmental stage-dependent effects of cluster bagging on phenolic metabolism in Cabernet Sauvignon grape berries. BMC Plant Biol 19:583. https://doi.org/10.1186/s12870-019-2186-z
Sunil L, Shetty NP (2022) Biosynthesis and regulation of anthocyanin pathway genes. Appl Microbiol Biotechnol 106:1783–1798. https://doi.org/10.1007/s00253-022-11835-z
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749. https://doi.org/10.1111/j.1365-313X.2008.03447.x
Tuan PA, Bai S, Yaegaki H, Tamura T, Hihara S, Moriguchi T, Oda K (2015) The crucial role of PpMYB10.1 in anthocyanin accumulation in peach and relationships between its allelic type and skin color phenotype. BMC Plant Biol 15:280. https://doi.org/10.1186/s12870-015-0664-5
Uematsu C, Katayama H, Makino I, Inagaki A, Arakawa O, Martin C (2014) Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. J Exp Bot 65:1081–1094. https://doi.org/10.1093/jxb/ert456
Wang J, Cao K, Wang LR, Dong WX, Zhang X, Liu WS (2022) Two MYB and three bHLH family genes participate in anthocyanin accumulation in the flesh of peach fruit treated with glucose, sucrose, sorbitol, and fructose in vitro. Plants-Basel 11. https://doi.org/10.3390/plants11040507
Wang QY, Zhu J, Li B, Li SS, Yang Y, Wang QY, Xu WZ, Wang LS (2023) Functional identification of anthocyanin glucosyltransferase genes: a Ps3GT catalyzes pelargonidin to pelargonidin 3-O-glucoside painting the vivid red flower color of Paeonia. Planta 257. https://doi.org/10.1007/s00425-023-04095-2
Xu W, Peng H, Yang T, Whitaker B, Huang L, Sun J, Chen P (2014) Effect of calcium on strawberry fruit flavonoid pathway gene expression and anthocyanin accumulation. Plant Physiol Biochem 82:289–298. https://doi.org/10.1016/j.plaphy.2014.06.015
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185. https://doi.org/10.1016/j.tplants.2014.12.001
Yan HL, Pei XN, Zhang H, Li X, Zhang XX, Zhao MH, Chiang VL, Sederoff RR, Zhao XY (2021) MYB-mediated regulation of anthocyanin biosynthesis. Int J Mol Sci 22. https://doi.org/10.3390/ijms22063103
Ying H, Shi J, Zhang S, Pingcuo G, Wang S, Zhao F, Cui Y, Zeng X (2019) Transcriptomic and metabolomic profiling provide novel insights into fruit development and flesh coloration in Prunus Mira Koehne, a special wild peach species. BMC Plant Biol 19:463. https://doi.org/10.1186/s12870-019-2074-6
Zhao X, Zhang W, Yin X, Su M, Sun C, Li X, Chen K (2015) Phenolic composition and antioxidant properties of different peach [Prunus persica (L.) Batsch] cultivars in China. Int J Mol Sci 16:5762–5778. https://doi.org/10.3390/ijms16035762
Zhao XY, Qi CH, Jiang H, You CX, Guan QM, Ma FW, Li YY, Hao YJ (2019) The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic Res 6:66. https://doi.org/10.1038/s41438-019-0147-1
Zhou Y, Zhou H, Lin-Wang K, Vimolmangkang S, Espley RV, Wang L, Allan AC, Han Y (2014) Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol 14:388. https://doi.org/10.1186/s12870-014-0388-y
Zhou H, Kui LW, Wang HL, Gu C, Dare AP, Espley RV, He HP, Allan AC, Han YP (2015) Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J 82:105–121. https://doi.org/10.1111/tpj.12792
Zhou H, Peng Q, Zhao JB, Owiti A, Ren F, Liao L, Wang L, Deng XB, Jiang Q, Han YP (2016) Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.01557
Acknowledgements
We thank Bioeditas Technology Corporation (Shaanxi, China) for helping with the transcriptome sequencing analysis and Metware Company for helping with the metabolome analysis.
Funding
This study was financially supported by the National Natural Science Foundation of China (32302508), the Natural Science Foundation of Shandong Province (No. ZR2021QC203), the Qilu University of Technology talent research project(2023RCKY222), the Basic Research Project of Science, the Education and Industry Integration Pilot Project (2022PY064), Key Innovation Project (2022JBZ01-06) of Qilu University of Technology (Shandong Academy of Sciences), and Development Plan of Youth Innovation Team in Colleges and Universities of Shandong Province 2022KJ127.
Author information
Authors and Affiliations
Contributions
methodology, XZ; validation, ZL, MC, and YY; formal analysis, ZL, SW, and MS; resources, XX; data curation, CX; writing—original draft preparation, CX; writing—review and editing, XZ; supervision, XZ; project administration, LQ; funding acquisition, WH. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, C., Xue, X., Li, Z. et al. The PpMYB75-PpDFR module reveals the difference between ‘SR’ and its bud variant ‘RMHC’ in peach red flesh. J Plant Res 137, 241–254 (2024). https://doi.org/10.1007/s10265-023-01512-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-023-01512-1