Abstract
Aluminum toxicity is the main factor limiting the elongation of plant roots in acidic soil. The tree species Eucalyptus camaldulensis is considerably more resistant to aluminum than herbaceous model plants and crops. Hydrolyzable tannins (HTs) accumulating in E. camaldulensis roots can bind and detoxify the aluminum taken up by the roots. However, in herbaceous model plants, HTs do not accumulate and the genes involved in the HT biosynthetic pathway are largely unknown. The aim of this study was to establish a method for reconstituting the HT biosynthetic pathway in the HT non-accumulating model plant Nicotiana benthamiana. Four E. camaldulensis enzymes were transiently expressed in N. benthamiana leaves via Agrobacterium tumefaciens-mediated transformation. These enzymes included dehydroquinate dehydratase/shikimate dehydrogenases (EcDQD/SDH2 and EcDQD/SDH3), which catalyze the synthesis of gallic acid, the first intermediate of the HT biosynthetic pathway that branches off from the shikimate pathway. The others were UDP-glycosyltransferases (UGT84A25 and UGT84A26), which catalyze the conversion of gallic acid to β-glucogallin, the second intermediate. The co-expression of the EcDQD/SDHs in transgenic N. benthamiana leaf regions promoted the synthesis of gallic acid. Moreover, the co-expression of the UGT84As in addition to the EcDQD/SDHs resulted in the biosynthesis of β-glucogallin, the universal metabolic precursor of HTs. Thus, we successfully reconstituted a portion of the HT biosynthetic pathway in HT non-accumulating N. benthamiana plants. This heterologous gene expression system will be useful for co-expressing candidate genes involved in downstream reactions in the HT biosynthetic pathway and for clarifying their in planta functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum (Al) toxicity is a major abiotic stress limiting the productivity of plants growing in acidic soil, which covers approximately 30% of the total land area worldwide (von Uexküll and Mutert 1995). Under acidic conditions below pH 5, Al is released into the soil solution from minerals, mainly as Al3+, and the accumulation of Al in root tips rapidly inhibits root elongation, ultimately leading to decreased water and nutrient uptake (Kopittke et al. 2016; Ma 2007). Therefore, the Al resistance of crops and trees must be improved to ensure sustainable food and forest production. Eucalyptus camaldulensis is a tree species that can grow in acidic soil and show no inhibition of root elongation when exposed to millimolar levels of Al (Tahara et al. 2008). The Al resistance of E. camaldulensis is 200- to 1,000-times higher than that of herbaceous model plants and crops. We previously isolated a hydrolyzable tannin (HT), oenothein B, from E. camaldulensis roots as a novel Al-binding ligand, and determined that the binding of Al3+ by oenothein B contributes to the detoxification of Al entering E. camaldulensis roots (Tahara et al. 2014). Tannins are a mixture of polyphenols found in plant leaves, bark, and wood, with molecular weights ranging from 500 to more than 3,000; they are divided into two types according to their chemical structure and properties: HTs and condensed tannins (Hagerman and Buttler 1981; Hassanpour et al. 2011). Condensed tannins are present in many plant species, whereas HTs are considered to accumulate preferentially in angiosperm plants excluding monocots (Bate-Smith 1984). Hydrolyzable tannins are widely distributed among long-lived woody plants, but they are absent in herbaceous model plants. This has hindered the elucidation of HT biosynthesis and regulation. Additionally, HTs reportedly have diverse functions, including as defense-related compounds that protect plants from herbivorous insects (Agrawal et al. 2012; Barbehenn and Constabel 2011) and mammals (Takahashi and Shimada 2008) as well as from microbial pathogens (Buzzini et al. 2008). Moreover, our previous research on E. camaldulensis showed that HTs have ecologically important effects because they can detoxify toxic metals (Tahara et al. 2014; 2017; Zhang et al. 2016). To effectively exploit these biological functions, the HT biosynthetic genes must be identified.
The HT biosynthetic pathway branches off from the shikimate pathway (Fig. 1). The shikimate pathway produces chorismate, which is an amino acid precursor, and is universally conserved in plants and microorganisms. In plants, shikimate dehydrogenase (SDH) family proteins may catalyze various reactions and link the shikimate pathway to HT and chlorogenic acid biosynthetic pathways (Bontpart et al. 2016; Guo et al. 2014). Plant SDHs form bifunctional enzymes by fusing with dehydroquinate dehydratases (DQDs), which catalyze the dehydration of 3-dehydroquinic acid to form 3-dehydroshikimic acid in the shikimate pathway (Peek and Christendat 2015). We identified four DQD/SDH family proteins (EcDQD/SDH1, 2, 3, and 4) in E. camaldulensis via heterologous protein production in Escherichia coli and in vitro catalytic activity assays (Fig. 2, Table S1). Notably, EcDQD/SDH2 and EcDQD/SDH3 catalyze the oxidation of 3-dehydroshikimic acid to gallic acid, which may link the shikimate pathway to HT biosynthesis (Fig. 1; Tahara et al. 2021). In contrast, EcDQD/SDH1 is responsible for the required reversible SDH activity in the shikimate pathway, while EcDQD/SDH4 exhibits reversible quinate dehydrogenase activity, which may link the shikimate pathway to chlorogenic acid biosynthesis. Arabidopsis thaliana has one DQD/SDH, which is more homologous to EcDQD/SDH1 than to the other DQD/SDHs in E. camaldulensis. Nicotiana benthamiana has seven DQD/SDHs that are homologous to EcDQD/SDH1 or 4 (Fig. 2). Thus, DQD/SDHs homologous to EcDQD/SDH2 and 3 are absent in these herbaceous model plants. Moreover, we previously identified the UDP-glycosyltransferases (UGTs) in E. camaldulensis (UGT84A25 and UGT84A26), which catalyze the conversion of gallic acid to β-glucogallin (1-O-galloyl-β-d-glucose; Figs. 1 and S1, Table S2; Tahara et al. 2018). However, the catalytic activities of these DQD/SDHs and UGTs in E. camaldulensis have been demonstrated by in vitro assays, but not in planta.
Overview of hydrolyzable tannin (HT), shikimate, and quinate biosynthesis in E. camaldulensis. Gallic acid biosynthesis is catalyzed by DQD/SDH family enzymes. Plant DQD/SDHs are proposed to link the shikimate pathway to the HT biosynthetic pathway. DQD dehydroquinate dehydratase, SDH shikimate dehydrogenase, UGT UDP-glycosyltransferase
Phylogenetic analysis of Nicotiana benthamiana DQD/SDHs and other functionally characterized DQD/SDH family members in seed plants. The phylogenetic tree was constructed based on an alignment of multiple full-length protein sequences according to the neighbor-joining method. The scale bar represents 0.1 fixed mutations per site. Bootstrap values (1,000 replicates) greater than 60% are indicated. The DQD/SDH family members can be classified into three groups whose SDH-domains mainly exhibit the activity of reversible shikimate dehydrogenase (SDH), gallic acid formation, or reversible quinate dehydrogenase (QDH). The accession numbers of the DQD/SDHs are listed in Table S1. The DQD/SDHs from N. benthamiana and Eucalyptus camaldulensis are highlighted in red and blue letters, respectively. Magenta dots indicate enzymes with gallate formation activity
There is currently no report regarding the biotechnological production of β-glucogallin in HT non-accumulating plants. Accordingly, the objective of this study was to reconstitute the relevant pathway in HT non-accumulating plants. We herein describe the Agrobacterium tumefaciens-mediated transient expression of the E. camaldulensis genes EcDQD/SDH2, EcDQD/SDH3, UGT84A25, and UGT84A26 in N. benthamiana leaves. Because of the similarities between N. benthamiana and other plants in terms of their cellular compartmentalization, cofactors, and coenzymes, there has been an increase in the use of N. benthamiana as a system for the reconstitution of the pathways associated with the production of plant natural compounds over the past decade (Reed and Osbourn 2018). The A. tumefaciens-mediated expression of EcDQD/SDH2 and EcDQD/SDH3 indicated that transgenic N. benthamiana leaf regions synthesized gallic acid. Furthermore, the co-expression of these four genes resulted in the production of β-glucogallin, the universal metabolic precursor of HTs. Therefore, the method established in this study may accelerate the characterization of the HT biosynthetic pathway and clarify HT functions in planta.
Materials and methods
Plant materials and growth conditions
Nicotiana benthamiana Domin plants were grown in a climate chamber under fluorescent light (16-h light/8-h dark) at 25 °C. A Eucalyptus camaldulensis Dehnh. clone (Myrtaceae; seed lot 19708; Australian Tree Seed Centre, CSIRO) was cultured hydroponically in a growth chamber as previously described (Tahara et al. 2018). Leaf samples were collected, immediately frozen in liquid nitrogen, and stored at − 80 °C until analyzed.
Phylogenetic analysis
The DQD/SDH and UGT sequences of N. benthamiana were obtained from the draft genome v1.01 (https://solgenomics.net/). After alignment with MUSCLE, the phylogenetic analyses of DQD/SDHs and UGTs sequences were conducted with the neighbor-joining method using MEGA11 (Tamura et al. 2021).
Taxonomic re-classification of HT distribution
Families and genera with HT-containing species determined on the basis of the Cronquist system of classification (Engelhardt et al. 2016; Okuda et al. 2000; Table S3, Fig. S2) were revised according to the Angiosperm Phylogeny Group classification (APG IV 2016).
RNA extraction
Total RNA was extracted from E. camaldulensis and N. benthamiana leaves using the hexadecyltrimethylammonium bromide (CTAB) method and purified using the SV Total RNA Isolation System (Promega) as previously described (Tahara et al. 2018).
Plasmid construction
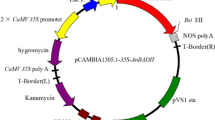
First-strand cDNA was synthesized from the total RNA extracted from E. camaldulensis leaves and then used as the template to amplify target cDNA sequences by PCR with specific primer sets (Table S4). For transient expression of multiple proteins simultaneously in leaves of N. benthamiana, we employed a polycistronic expression system using intein-UBQ sequence (Zhang et al. 2019) to minimize the use of the same promoter and to achieve equal expression levels of the related enzyme genes. For the biosynthesis of gallic acid, the intein-UBQ sequence was sandwiched by the open reading frames of EcDQD/SDH2 (Accession No. LC487989) and EcDQD/SDH3 (LC487990) and subsequently inserted into p35SHSPstarG4_L4R1, which is the same as p35SHSPG (Oshima et al. 2011) except for the mutated HindIII site in the HSP terminator and the different Gateway att site (attL4–attR1) (Fig. 3a). The same method was used to prepare the construct for UGT84A25 (LC189069) and UGT84A26 (LC189071), which are responsible for the subsequent reaction, but the construct was inserted into p35SHSPG, which contains the Gateway attL1–attL2 site (Oshima et al. 2011; Fig. 3b). Finally, the inserted constructs in these two plasmids and the corresponding sequence in the empty vector were inserted into the pGWB501 vector (Nakagawa et al. 2009), which has the Gateway attR4–attR2 site, via a multisite Gateway reaction to prepare the recombinant plasmids for the transient expression of SDH, UGT, and SDH-UGT.
Schematic representation of the expression vectors used for the transient expression in N. benthamiana. a SDH plasmid: the cauliflower mosaic virus (CaMV) 35S promoter was fused to the EcDQD/SDH2 and EcDQD/SDH3 genes that sandwiched the intein-UBQ (IU) sequence. b UGT plasmid: the CaMV 35S promoter was fused to the UGT84A25 and UGT84A26 genes that sandwiched the IU sequence. c SDH-UGT plasmid: the expression cassettes from the SDH and UGT plasmids were ligated in tandem. After translation, the polyprotein precursor is cleaved at the N- and C-termini of the IU domain, resulting in the release of two separate proteins (Zhang et al. 2019). HSPt, heat shock protein terminator of Arabidopsis thaliana
Transient heterologous gene expression in N. benthamiana
The recombinant plasmids were individually introduced into Agrobacterium tumefaciens strain GV3101 cells harboring the transformation helper plasmid pSoup via electroporation (Hellens et al. 2000).
Overnight cultures were harvested by centrifugation and the pellets were resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES pH 5.6, and 100 μM acetosyringone). The OD600 values of the suspensions were adjusted to 0.5, after which the suspensions were incubated for 2 h at room temperature prior to the infiltration of N. benthamiana using a needleless syringe. The infiltrated plants were cultivated in the climate chamber. At specific time-points, leaves were collected and immediately frozen in liquid nitrogen.
Quantitative real-time PCR (RT-qPCR) analysis
PrimeScript RT Master Mix (Takara) was used to synthesize cDNA from the extracted total RNA. The RT-qPCR analysis of EcDQD/SDH2, EcDQD/SDH3, UGT84A25, and UGT84A26 transcript levels was performed using TB Green Premix Ex Taq II (Takara) and the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The N. benthamiana gene encoding elongation factor 1-α (NbEF-1α) transcripts served as internal control. The gene-specific primers are listed in Table S4. The target sequences were amplified using the E. camaldulensis cDNA as the template and then cloned into a pBlueScript II SK ( +) vector. The NbEF-1α was amplified using the N. benthamiana cDNA as the template and then cloned into a p35SHSPG vector (Oshima et al. 2011). The plasmid solution for each gene was serially diluted tenfold (from 108 to 103 molecules μL−1) and used to generate a standard curve for the absolute quantification. To normalize against the NbEF-1α transcript, the relative mRNA copy number of EcDQD/SDH2, EcDQD/SDH3, UGT84A25, and UGT84A26 was calculated by the following equation:
Identification and quantification of HTs and related compounds
Leaf samples (1 g) were ground to a powder in liquid nitrogen using a cell disruptor (Multi-Beads Shocker, Yasui Kikai) for the subsequent extraction with 1 mL ice-cold 70% (v/v) aqueous acetone. The samples were centrifuged at 4 °C and the supernatant (50 µL) was diluted with 450 µL 0.1% (v/v) formic acid in water. Extracts were analyzed using an ultra-performance liquid chromatography (UPLC) system (ACQUITY UPLC H-Class, Waters) coupled with a quadrupole time-of-flight mass spectrometry (Q-TOF–MS) system (Xevo G2-XS QTof, Waters). The UPLC conditions were as follows: column, ACQUITY UPLC HSS T3 Column (particle size 1.8 µm, 2.1 mm × 100 mm; Waters); column temperature, 40 °C; solvent A, 0.1% (v/v) formic acid in water; solvent B, acetonitrile; flow rate, 0.5 mL min−1. The gradient was 0.1–25% eluent B (at 0–10 min after injection), 25–99% (10–10.1 min), 99–0.1% (12–12.1 min), and 0.1% (12.1–15 min). The Q-TOF–MS conditions were as follows: ionization mode, electrospray ionization; acquisition mode, MSE or MS/MS negative-sensitivity mode; acquisition range, 40–1,200; capillary voltage, 1.0 kV; cone voltage, 30 V; source temperature, 120 °C; desolvation temperature, 500 °C; cone gas flow, 50 L h−1; desolvation gas flow, 1,000 L h−1; collision energy, 6 eV (low energy) or 10–45 eV (high energy). Compounds in the extracts were identified by comparing their retention times and product ion spectra with those of authentic standard compounds (β-glucogallin, BOC Sciences; 3-glucogallic acid and 4-glucogallic acid, Synthose). The measured mass of the compounds was consistent with their elemental composition (Table S5). Each compound was quantified according to a standard curve plotted using the peak areas in the extracted ion chromatograms of the deprotonated molecule m/z values.
Extraction of crude protein from plants and UGT activity assay
Frozen fresh leaves (0.5 g) were mixed with 2 mL ice-cold extraction buffer (100 mM phosphate buffer pH 7.5 and 150 mM NaCl) supplemented with 0.5 g PVPP and then homogenized in a mortar. The homogenized samples were centrifuged. The supernatant was used for the overnight precipitation with ammonium sulfate (80% saturation) at 4 °C. The precipitate was suspended in 1 mL storage buffer [100 mM MES pH 5.5, 100 mM NaCl, and 10% (v/v) glycerol]. After a centrifugation at 10,000×g for 5 min, the supernatant was desalted using the Amicon Ultra-0.5 Centrifugal Filter Device (Merck Millipore). The protein concentrations of desalted extract were estimated using the Qubit Protein Assay Kit (Thermo Fisher Scientific) prior to the UGT activity assay.
The UGT activity assay was performed using a 100 μL reaction mixture containing 100 mM MES (pH 5.5), 4 mM UDP-glucose, 2 mM gallic acid, 3 mM 2-mercaptoethanol, and 50 μg desalted crude protein. The reaction mixture was incubated at 30 °C for 3 h. All reactions were terminated by adding an equal volume of methanol and then samples were centrifuged. The supernatant was transferred to a new tube for the UPLC-Q-TOF–MS analysis.
Statistical analysis
The metabolite concentrations and gene expression levels are presented herein as the mean ± SE for at least three replicates. Data were analyzed using Student’s t-test or the Tukey–Kramer test with BellCurve for Excel version 4.04 (Social Survey Research Information).
Results
Distribution of HTs among flowering plants
Hydrolyzable tannins accumulate in angiosperms, but not in gymnosperms. The taxonomic classification of HT-containing plants based on the Cronquist system of classification (Okuda et al. 2000) was revised according to the Angiosperm Phylogeny Group classification (APG IV 2016, Fig. S2). The orders, families, and genera in which HTs have been detected are listed in Table S3. Most of the taxonomic orders in which HTs have been detected were revealed to comprise a particular group of angiosperms (i.e., core eudicots). Eucalyptus camaldulensis from the family Myrtaceae belongs to this group. Although A. thaliana (i.e., herbaceous model plant) also belongs to this group, there are no reports indicating it accumulates HTs. The herbaceous model plant used in this study, N. benthamiana, belongs to a different group (Lamiids), which does not include plant species that accumulate HTs. Exceptionally, HTs reportedly accumulate in Nuphar japonicum, which belongs to the order Nymphaeales, the second plant group that separated from the others after the order Amborellales.
Transient co-expression of E. camaldulensis DQD/SDH and UGT transgenes in N. benthamiana leaves
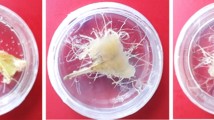
During our earlier research on E. camaldulensis, we identified EcDQD/SDH2 and EcDQD/SDH3, which catalyze the formation of gallic acid, and UGT84A25 and UGT84A26, which catalyze the synthesis of β-glucogallin. In a specific branch of the shikimate pathway, the gallic acid produced by EcDQD/SDH2 and EcDQD/SDH3 may be further metabolized by UGT84A25 and UGT84A26 (Fig. 1). To express multiple proteins simultaneously, we constructed recombinant plasmids containing EcDQD/SDH2 and 3 and/or UGT84A25 and 26 under the control of the 35S promoter by employing the polycistronic expression system using intein-UBQ (IU) sequence (Fig. 3). The plasmids (SDH, UGT, and SDH-UGT) were then introduced into A. tumefaciens strain GV3101 via electroporation. First, a 5-day transgene expression analysis was performed to determine the appropriate time for sampling. Agrobacterium tumefaciens cells harboring the empty vector or SDH-UGT vector were used for the agroinfiltration of N. benthamiana leaves, which were then collected at specific time-points for the analysis of transgene expression by RT-qPCR (Fig. 4). Transgene expression was undetectable in the leaves infiltrated with A. tumefaciens cells containing the empty vector. The time-course analysis of the leaves infiltrated with A. tumefaciens cells containing the SDH-UGT vector revealed a lack of significant differences in transgene expression at 3–5 days post-infiltration (Fig. 4b). Moreover, leaves expressing SDH-UGT were pale green and yellow at 4 and 5 days post-infiltration, respectively (Fig. 4a). They were mostly necrotic at 6 days post-infiltration (Fig. S3). Thus, 3 days post-infiltration was selected as the sampling time-point.
Determination of the optimal sample collection time according to a time-course analysis of transgene expression. a The N. benthamiana plants infiltrated with A. tumefaciens harboring the empty vector or the SDH-UGT vector are presented. The arrows indicate pale green leaves at 4 days and yellow leaves at 5 days post-infiltration. b RT-qPCR analyses were performed using gene-specific primers and separate standard curves. The mRNA copy numbers were normalized against the mRNA copy number of the gene encoding elongation factor 1-α (NbEF-1α). Data are presented as the mean ± SE (n = 3). ND not detectable. Identical letters indicate a lack of significant difference at P < 0.05 (Tukey–Kramer test)
Expression of E. camaldulensis DQD/SDH and UGT transgenes enables the biosynthesis of the HT precursor in N. benthamiana
Agrobacterium tumefaciens cells harboring the empty control vector or the SDH, UGT, and SDH-UGT expression vectors were used for the infiltration of N. benthamiana leaves. Transgene expression levels in the leaves collected 3 days later were quantified by RT-qPCR and were recorded as mRNA copy numbers relative to the NbEF-1α mRNA copy number (Fig. 5). Both EcDQD/SDH2 and EcDQD/SDH3 were expressed in the leaves infiltrated with A. tumefaciens harboring SDH and SDH-UGT. Similarly, UGT84A25 and UGT84A26 were expressed in the leaves infiltrated with A. tumefaciens harboring UGT and SDH-UGT.
Transgene expression in leaves infiltrated with A. tumefaciens harboring the empty, SDH, UGT, or SDH-UGT vector. Total RNA was extracted from the leaves collected 3 days post-infiltration for the RT-qPCR analyses performed using gene-specific primers and separate standard curves. The mRNA copy numbers were normalized against the mRNA copy number of the gene encoding elongation factor 1-α (NbEF-1α). Data are presented as the mean ± SE (n = 3). ND, not detectable. Asterisks indicate a significant difference at **P < 0.01 (Student’s t-test) in the EcDQD/SDH2 and 3 expression levels between the leaves expressing SDH and the leaves expressing SDH-UGT. Asterisks indicate a difference at **P < 0.01 (Student’s t-test) in the UGT84A25 and 26 expression levels between the leaves expressing UGT and the leaves expressing SDH-UGT
To confirm DQD/SDH and UGT genes contribute to HT biosynthesis in planta, metabolites were extracted from agroinfiltrated leaves and then analyzed by UPLC-Q-TOF–MS. The extracted ion chromatograms (m/z 169.01) for the leaves expressing SDH and SDH-UGT had an obvious peak with the same retention time as gallic acid (Fig. 6a). Furthermore, the product ion spectrum of the peak revealed a fragmentation pattern that was identical to that of gallic acid (Fig. S4). Approximately 0.3–1 nmol gallic acid was detected in 1 g leaves expressing SDH and SDH-UGT, whereas gallic acid was undetectable in the leaves infiltrated with A. tumefaciens harboring UGT (Fig. 6b). Unexpectedly, a very small amount of gallic acid was detected in the leaves infiltrated with buffer or A. tumefaciens harboring the empty vector. The extracted ion chromatograms (m/z 331.07) for the leaves expressing SDH-UGT contained an obvious peak with the same retention time as β-glucogallin (Fig. 6a). The product ion spectrum of the peak had a fragmentation pattern identical to that of β-glucogallin (Fig. S4). Approximately 10 nmol β-glucogallin was detected in 1 g leaves expressing SDH-UGT, but β-glucogallin was not detected in the leaves infiltrated with A. tumefaciens harboring the SDH vector or in the control leaves (Fig. 6b). The heterologous expression experiments were repeated more than 10 times, with similar results each time. The generated data suggest that the heterologous expression of EcDQD/SDH2 and 3 as well as UGT84A25 and 26 enables the biosynthesis of HT precursors (gallic acid and β-glucogallin) in the HT non-accumulating herbaceous model plant N. benthamiana. The concentrations of gallic acid and β-glucogallin produced in N. benthamiana were roughly one-hundredth of those in E. camaldulensis. In 1 g leaves of E. camaldulensis, 63 ± 14 nmol gallic acid and 420 ± 180 nmol β-glucogallin were detected (mean ± SD).
Formation of gallic acid, β-glucogallin, 3-glucogallic acid and 4-glucogallic acid in N. benthamiana leaves transiently expressing E. camaldulensis DQD/SDH and/or UGT transgenes. Leaves were collected at 3 days post-infiltration with infiltration buffer alone or A. tumefaciens harboring the empty, SDH, UGT, or SDH-UGT vector. a The UPLC-Q-TOF–MS peak profiles revealed gallic acid was formed in the leaves expressing SDH and SDH-UGT, while β-glucogallin was formed in the leaves expressing SDH-UGT. b Concentrations of gallic acid, β-glucogallin, 3-glucogallic acid and 4-glucogallic acid biosynthesized in transgenic N. benthamiana leaf regions. Data are presented as the mean ± SE (n = 3–7). ND, not detectable. Identical letters indicate a lack of significant difference at P < 0.05 (Tukey–Kramer test)
Endogenous UGTs in N. benthamiana glucosylate gallic acid to form 3-glucogallic acid and 4-glucogallic acid
Interestingly, the extracted ion chromatograms (m/z 331.07) for the leaves expressing SDH and SDH-UGT had two obvious peaks at 2.6 and 3.8 min (Fig. 6a), implying the expression of SDH and SDH-UGT promoted the accumulation of two metabolites other than gallic acid and β-glucogallin. The deprotonated molecule peak (m/z 331.07) and the product ion peak (m/z 169.01) in the mass spectra (Fig. S4) suggested that the two metabolites have the same molecular weight as β-glucogallin and contain gallic acid as a substructure. We identified them as 3-glucogallic acid (gallic acid 3-O-β-D-glucoside) and 4-glucogallic acid (gallic acid 4-O-β-d-glucoside) by comparing their retention times and product ion spectra with those of authentic standards (Figs. 6a, 7 and S4). Approximately 10 nmol 3-glucogallic acid and 2 nmol 4-glucogallic acid were detected in 1 g leaves expressing SDH. In addition, a small amount of 3-glucogallic acid was detected in all examined leaves (Fig. 6b). These results suggest that endogenous glucosyltransferases in N. benthamiana catalyze the conversion of gallic acid to 3-glucogallic acid or 4-glucogallic acid.
A model biosynthetic pathway for β-glucogallin, 3-glucogallic acid and 4-glucogallic acid in N. benthamiana leaves transiently expressing E. camaldulensis UGT84As. While exogenous E. camaldulensis UGT84A25 and UGT84A26 catalyze the formation of β-glucogallin, endogenous N. benthamiana UGTs catalyze the formation of 3-glucogallic acid and 4-glucogallic acid. The position of the hydroxy group of gallic acid, to which glucose is transferred by UGT, determines which gallic acid derivative is formed
To verify this possibility, we infiltrated N. benthamiana leaves with buffer or A. tumefaciens harboring the empty vector or the UGT expression vector. The crude protein extracts obtained from the N. benthamiana leaves were used for in vitro catalytic assays, with gallic acid and UDP-glucose as substrates. The reaction products were analyzed by UPLC-Q-TOF–MS, which revealed that the native crude proteins from N. benthamiana leaves produced 3-glucogallic acid and 4-glucogallic acid. In addition, only proteins from the N. benthamiana leaves expressing UGTs produced β-glucogallin (Fig. 8). All extracted ion chromatograms for the denatured proteins used in assays lacked obvious peaks. These results suggest that endogenous UDP-glucose-dependent glucosyltransferases in N. benthamiana catalyze the transfer of glucose to gallic acid to form 3-glucogallic acid or 4-glucogallic acid, but not β-glucogallin (Fig. 7).
In vitro functional analysis of E. camaldulensis UGTs and endogenous N. benthamiana UGTs. Crude protein extracts from N. benthamiana leaves infiltrated with infiltration buffer alone or with A. tumefaciens harboring the empty or UGT vector were used to assay in vitro enzyme activities, with gallic acid and UDP-glucose as substrates. The UPLC-Q-TOF–MS peak profiles indicated 3-glucogallic acid and 4-glucogallic acid were formed in all assays involving native proteins. Additionally, β-glucogallin was formed in the assay involving native proteins from the leaves expressing UGT. The results of the enzyme activity assay using the native and denatured proteins are presented as peak profiles with orange and blue lines, respectively
Effects of the transient expression of E. camaldulensis SDH and UGT transgenes on other metabolites
To clarify how the transient expression of E. camaldulensis DQD/SDH and UGT transgenes affects other metabolites related to the shikimate pathway, metabolites extracted from agroinfiltrated leaves were analyzed by UPLC-Q-TOF–MS (Fig. 9). The transient expression of SDH, UGT, or SDH-UGT had no significant effect on the 3-dehydroshikimic acid and quinic acid concentrations. In contrast, the shikimic acid concentration increased in the leaves in which SDH was transiently expressed. The 3-dehydroquinic acid concentration was too low to quantify. These findings indicate that the yellowing of the leaves agroinfiltrated with SDH-UGT was probably due to the increased synthesis of HT precursors rather than a decrease in native metabolite contents.
Effect of the heterologous expression of E. camaldulensis SDH and UGT transgenes on shikimic acid and quinic acid biosynthesis. The concentrations of quinic acid, 3-dehydroshikimic acid, and shikimic acid in the transgenic N. benthamiana leaf regions in Fig. 6 are indicated. Data are presented as the mean ± SE (n = 3–7). Identical letters indicate a lack of significant difference at P < 0.05 (Tukey–Kramer test)
Discussion
Demonstration of the E. camaldulensis DQD/SDH and UGT functions in planta
With the ultimate goal of enhancing the Al resistance of crops and trees, we attempted to identify and characterize the E. camaldulensis genes involved in HT biosynthesis. The heterologous protein production in E. coli and in vitro catalytic assays with purified enzymes showed that EcDQD/SDH2 and 3 catalyze the synthesis of gallic acid, the first intermediate in the HT biosynthetic pathway (Tahara et al. 2021). Moreover, the heterologous protein production in E. coli and in vitro assays demonstrated that UGT84A25 and 26 catalyze the esterification of UDP-glucose and gallic acid to form β-glucogallin, the second HT intermediate (Tahara et al. 2018). In vitro assays indicated that these enzymes (DQD/SDHs and UGTs) are important for the biosynthesis of HT precursors, but their functions in planta will need to be confirmed. N. benthamiana does not accumulate HTs, but pathways for several natural products, including betalains (Polturak et al. 2016) and terpenoids (Reed and Osbourn 2018), have been successfully reconstituted in this herbaceous model plant via Agrobacterium-mediated transient expression. In the current study, the biosynthesis of gallic acid in N. benthamiana leaves transiently expressing EcDQD/SDH2 and EcDQD/SDH3 reflected the catalytic activity of E. camaldulensis DQD/SDHs in planta (Figs. 6 and S4). In addition, the A. tumefaciens-mediated co-expression of E. camaldulensis UGT84A25 and UGT84A26 with DQD/SDHs resulted in the production of β-glucogallin in the transgenic N. benthamiana leaf regions (Figs. 6 and S4). Thus, E. camaldulensis UGTs can catalyze the biosynthesis of β-glucogallin in planta, with the gallic acid produced by SDH serving as a substrate.
Successful reconstitution of part of the HT biosynthetic pathway in HT non-accumulating plants
The DQD/SDHs that catalyze the oxidation of 3-dehydroshikimic acid to produce gallic acid have also been identified in Juglans regia (Muir et al. 2011) and Vitis vinifera (Bontpart et al. 2016). Additionally, J. regia DQD/SDH was heterologously overexpressed in N. tabacum, which resulted in increased gallic acid accumulation (Muir et al. 2011). There is currently no report regarding the biotechnology-based production of the subsequent intermediate β-glucogallin in HT non-accumulating plants. In this study, the A. tumefaciens-mediated transient overexpression of E. camaldulensis DQD/SDH and UGT transgenes led to the formation of β-glucogallin in HT non-accumulating plants. A time-course analysis of transgene expression showed that the leaves agroinfiltrated with SDH-UGT were wilted and necrotic at the infiltration site after a long-term incubation, whereas the control leaves were normal (Figs. 4a and S3). Our unpublished study showed that the addition of 16 μM β-glucogallin to the culture medium inhibited the root elongation of A. thaliana, indicating the phytotoxicity of β-glucogallin. These observations suggest that the continuous accumulation of β-glucogallin in HT non-accumulating plants may have toxic effects. This toxicity might explain why transformed plants have not been reported to date. Transformants may be obtained using inducible vectors.
Utility of the SDH-UGT expression system for functionally characterizing downstream candidate genes in planta
There has recently been rapid progress in the whole-genome sequencing of various plants because of the advancements in sequencing technologies, but the genetic mechanisms controlling the biosynthesis and accumulation of HTs remain relatively unknown (e.g., Wang et al. 2021). Specifically, although DQD/SDH and UGT84A family genes encoding enzymes that catalyze the synthesis of the first and second intermediates, respectively, have been identified, the genes encoding the enzymes involved in the formation of the third and higher-order intermediates remain to be identified. The third to sixth intermediates are galloylglucoses (i.e., 1,6-di-, to 1,2,3,4,6-penta-O-galloyl-β-d-glucoses), which are synthesized by a series of galloylations of galloylglucoses, with β-glucogallin as the galloyl donor (Niemetz and Gross 2005). Researchers speculated that serine carboxypeptidase-like acyltransferase (SCPL-AT) family proteins catalyze the galloylation reaction because the expression levels of some SCPL-AT homologs are consistent with the biosynthesis of galloylated flavan-3-ols in grape berries (V. vinifera; Bontpart et al. 2018) and persimmon fruits (Diospyros kaki; Akagi et al. 2009). However, nearly all of the SCPL-AT candidates will need to be functionally characterized to determine whether and how they are involved in the galloylation of metabolites in plants. Because most SCPL-ATs undergo complex post-translational processing and are secreted into organelles, it may be difficult to provide the functional proof of SCPL-ATs (Mugford and Milkowski 2012). Two SCPL-ATs (CsSCPL4 and CsSCPL5) involved in the galloylation of flavan-3-ols (epicatechin and epigallocatechin) were recently isolated from Camellia sinensis (Yao et al. 2022). The co-expression, post-transcriptional processing, and interactions of CsSCPL4 and CsSCPL5 in N. benthamiana, but not in E. coli, are required for their galloylation activities. In the current study, we developed a transient heterologous SDH-UGT expression system that allows HT non-accumulating N. benthamiana plants to accumulate β-glucogallin (i.e., HT precursor). This system may be useful for co-expressing candidate genes and elucidating the in planta functions of the encoded enzymes (e.g., SCPL-ATs) that may catalyze downstream reactions in the HT biosynthetic pathway. Hence, this system will accelerate the characterization of the HT biosynthetic pathway as well as the physiological functions of HTs in plants.
Effect of the partial reconstitution of the HT biosynthetic pathway on related metabolites
A small amount of gallic acid was biosynthesized in all N. benthamiana leaves, even in the wild-type leaves (Fig. 6). This is consistent with the fact that a small amount of β-glucogallin was produced in the leaves expressing UGT (Fig. 6). In the leaves expressing SDH, gallic acid as well as 3-glucogallic acid and 4-glucogallic acid levels increased (Fig. 6). This phenomenon is due to endogenous N. benthamiana UGTs that catalyze the reaction that converts gallic acid to 3-glucogallic acid or 4-glucogallic acid (Figs. 7 and 8). The endogenous UGTs likely metabolize gallic acid into 3-glucogallic acid, even in the wild-type N. benthamiana leaves. The glucosylation by endogenous UGTs helps protect plant cells from the deleterious effects of the reactive intermediate gallic acid. In the leaves expressing SDH-UGT, three types of gallic acid derivatives with a glucose were formed (Figs. 6 and 7). The type of gallic acid derivative is determined by the position of the hydroxy group of gallic acid, to which glucose is transferred by UGT (Fig. 7). Among the three derivatives, β-glucogallin (1-O-β-d-galloylglucose) is an energy-rich β-acetal ester that serves as a galloyl donor during galloylation reactions, whereas 3-glucogallic acid and 4-glucogallic acid are glucosides. Although the enzymes that catalyze the conversion of gallic acid to 3-glucogallic acid have not been identified in plants, UGT72BD1 reportedly catalyzes the reaction that converts gallic acid to 4-glucogallic acid in pomegranate (Punica granatum) (Chang et al. 2019). Similar to UGT72BD1, some members of the UGT72 family glucosylate phenylpropanoids at the C4 position of their aromatic ring, forming 4-O-glucosides (Fig. S1). In the genome sequence of N. benthamiana, there are nine UGT72 family genes (Fig. S1), which could potentially encode UGTs responsible for 4-glucogallic acid formation in N. benthamiana. A comprehensive database of plant UDP-dependent glycosyltransferases (pUGTdb) has been released and will be useful for functionally annotating these enzymes (Liu et al. 2023). In the present study, shikimic acid levels were higher in the leaves overexpressing DQD/SDH transgenes (e.g., SDH and SDH-UGT) than in the control leaves (Fig. 9). The increase in shikimic acid may be due to the fact that EcDQD/SDH2 and EcDQD/SDH3 have shikimate-forming activities in addition to their gallate-forming activities (Tahara et al. 2021).
Data availability
The data supporting the findings of this study are available on request from the corresponding author.
Change history
17 August 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10265-023-01488-y
References
Agrawal AA, Hastings AP, Johnson MTJ, Maron JL, Salminen JP (2012) Insect herbivores drive real-time ecological and evolutionary change in plant populations. Science 338:113–116. https://doi.org/10.1126/science.1225977
Akagi T, Ikegami A, Suzuki Y, Yoshida J, Yamada M, Sato A, Yonemori K (2009) Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kakiThunb.) fruit. Planta 230:899–915. https://doi.org/10.1007/s00425-009-0991-6
APG IV (2016) An update of the angiosperm phylogeny group classificaion for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181:1–20. https://doi.org/10.1111/boj.12385
Barbehenn RV, Constabel CP (2011) Tannins in plant-herbivore interactions. Phytochemistry 72:1551–1565. https://doi.org/10.1016/j.phytochem.2011.01.040
Bate-Smith EC (1984) Age and distribution of galloyl esters, iridoids and certain other repellents in plants. Phytochemistry 23:945–950. https://doi.org/10.1016/S0031-9422(00)82588-1
Bontpart T, Marlin T, Vialet S, Guiraud JL, Pinasseau L, Meudec E, Sommerer N, Cheynier V, Terrier N (2016) Two shikimate dehydrogenases, VvSDH3 and VvSDH4, are involved in gallic acid biosynthesis in grapevine. J Exp Bot 67:3537–3550. https://doi.org/10.1093/jxb/erw184
Bontpart T, Ferrero M, Khater F, Marlin T, Vialet S, Vallverdù-Queralt A, Pinasseau L, Ageorges A, Cheynier V, Terrier N (2018) Focus on putative serine carboxypeptidase-like acyl-transferases in grapevine. Plant Physiol Biochem 130:356–366. https://doi.org/10.1016/j.plaphy.2018.07.023
Buzzini P, Arapitsas P, Goretti M, Branda E, Turchetti B, Pinelli P, Ieri F, Romani A (2008) Antimicrobial and antiviral activity of hydrolysable tannins. Mini Rev Med Chem 8:1179–1187. https://doi.org/10.2174/138955708786140990
Chang L, Wu S, Tian L (2019) Effective genome editing and identification of a regiospecific gallic acid 4-O-glycosyltransferase in pomegranate (Punica granatum L.). Hort Res 6:123. https://doi.org/10.1038/s41438-019-0206-7
Engelhardt C, Petereit F, Lechtenberg M, Liefländer-Wulf U, Hensel A (2016) Qualitative and quantitative phytochemical characterization of Myrothamnus flabellifolia Welw. Fitoterapia 114:69–80. https://doi.org/10.1016/j.fitote.2016.08.013
Guo J, Carrington Y, Alber A, Ehlting J (2014) Molecular characterization of quinate and shikimate metabolism in Populus trichocarpa. J Biol Chem 289:23846–23858. https://doi.org/10.1074/jbc.M114.558536
Hagerman AE, Buttler LG (1981) The specificity of proanthocyanidin-protein interactions. J Bio Chem 256:4494–4497. https://doi.org/10.1016/S0021-9258(19)69462-7
Hassanpour S, Maheri-Sis N, Eshratkhah B, Mehmandar FB (2011) Plants and secondary metabolites (Tannins): a review. Int J For Soil Eros 1:47–53
Hellens RP, Edwards AE, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42:819–832
Kopittke PM, Menzies NW, Wang P, Blamey FPC (2016) Kinetics and nature of aluminium rhizotoxic effects: a review. J Exp Bot 67:4451–4467. https://doi.org/10.1093/jxb/erw233
Liu Y et al (2023) pUGTdb: a comprehensive database of plant UDP-dependent glycosyltransferases. Mol Plant 16:634–646. https://doi.org/10.1016/j.molp.2023.01.003
Ma JF (2007) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol 264:225–252. https://doi.org/10.1016/S0074-7696(07)64005-4
Mugford ST, Milkowski C (2012) Serine carboxypeptidase-like acyl-transferases from plants. In: Hopwood DA (ed) Natural product biosynthesis by microorganisms and plants, part B. Elsevier Academic Press Inc, San Diego, pp 279–297
Muir RM, Ibanez AM, Uratsu SL, Ingham ES, Leslie CA, McGranahan GH, Batra N, Goyal S, Joseph J, Jemmis ED, Dandekar AM (2011) Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol Biol 75:555–565. https://doi.org/10.1007/s11103-011-9739-3
Nakagawa T, Ishiguro S, Kimura T (2009) Gateway vectors for plant transformation. Plant Biotechnol 26:275–284. https://doi.org/10.5511/plantbiotechnology.26.275
Niemetz R, Gross GG (2005) Enzymology of gallotannin and ellagitannin biosynthesis. Phytochemistry 66:2001–2011. https://doi.org/10.1016/j.phytochem.2005.01.009
Okuda T, Yoshida T, Hatano T (2000) Correlation of oxidative transformations of hydrolysable tannins and plant evolution. Phytochemistry 55:513–529. https://doi.org/10.1016/S0031-9422(00)00232-6
Oshima Y, Mitsuda N, Nakata M, Nakagawa T, Nagaya S, Kato K, Ohme-Takagi M (2011) Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol 28:201–210. https://doi.org/10.5511/plantbiotechnology.11.0124a
Peek J, Christendat D (2015) The shikimate dehydrogenase family: functional diversity within a conserved structural and mechanistic framework. Arch Biochem Biophys 566:85–99. https://doi.org/10.1016/j.abb.2014.12.006
Polturak G, Breitel D, Grossman N, Sarrion-Perdigones A, Weithorn E, Pliner M, Orzaez D, Granell A, Rogachev I, Aharoni A (2016) Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol 210:269–283. https://doi.org/10.1111/nph.13796
Reed J, Osbourn A (2018) Engineering terpenoid production through transient expression in Nicotiana benthamiana. Plant Cell Rep 37:1431–1441. https://doi.org/10.1007/s00299-018-2296-3
Tahara K, Norisada M, Yamanoshita T, Kojima K (2008) Role of aluminum-binding ligands in aluminum resistance of Eucalyptus camaldulensis and Melaleuca cajuputi. Plant Soil 302:175–187. https://doi.org/10.1007/s11104-007-9464-5
Tahara K, Hashidada K, Otsuka Y, Ohara S, Kojima K, Shinohara K (2014) Identification of hydrolysable tannin, oenothein B, as an aluminum-detoxifying ligand in a highly aluminum-resistant tree, Eucalyptus camaldulensis. Plant Physiol 164:683–693. https://doi.org/10.1104/pp.113.222885
Tahara K, Hiradate S, Hashida K, Shinohara K (2017) An aluminum-resistance mechanism in Eucalyptus camaldulensis: complexation between aluminum and oenothein B in presence of organic acids in vitro. J for Res 22:261–264. https://doi.org/10.1080/13416979.2017.1326656
Tahara K, Nishiguchi M, Frolov A, Mittasch J, Milkowski C (2018) Identification of UDP glucosyltransferase from the aluminum-resistance tree Eucalyptus camaldulensis forming β-glucogallin, the precursor of hydrolyzable tannins. Phytochemistry 152:154–161. https://doi.org/10.1016/j.phytochem.2018.05.005
Tahara K, Nishiguchi M, Funke E, Miyazawa S, Miyama T, Milkowski C (2021) Dehydroquinate dehydratase/shikimate dehydrogenases involved in gallate biosynthesis of the aluminum-tolerant tree species Eucalyptus camaldulensis. Planta 253:3. https://doi.org/10.1007/s00425-020-03516-w
Takahashi A, Shimada T (2008) Selective consumption of acorns by the Japanese wood mouse according to tannin content: a behavioral countermeasure against plant secondary metabolites. Ecol Res 23:1033–1038. https://doi.org/10.1007/s11284-008-0473-5
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Von Uexküll HR, Mutert E (1995) Global extent, development and economic impact of acid soils. Plant Soil 171:1–15. https://doi.org/10.1007/BF00009558
Wang L et al (2021) The chromosome-scale reference genome of Rubus chingii Hu provides insight into the biosynthetic pathway of hydrolysable tannins. Plant J 107:1466–1477. https://doi.org/10.1111/tpj.15394
Yao S et al (2022) Insights into acylation mechanisms: co-expression of serine carboxypeptidase-like acyltransferases and their non-catalytic companion paralogs. Plant J 111:117–133. https://doi.org/10.1111/tpj.15782
Zhang L, Liu R, Gung BW, Tindall S, Gonzalez JM, Halvorson JJ, Hagerman AE (2016) Polyphenol-aluminum complex formation: implications for aluminum tolerance in plants. J Agric Food Chem 64:3025–3033. https://doi.org/10.1021/acs.jafc.6b00331
Zhang B, Han Z, Kumar S, Gupta M, Su WW (2019) Intein-ubiquitin chimeric domain for coordinated protein coexpression. J Biotechnol 304:38–43. https://doi.org/10.1016/j.jbiotec.2019.08.006
Acknowledgements
We appreciate the technical assistance provided by Ms. Emiko Aizawa (FFPRI). This study was supported by the Support Program of FFPRI for researchers having family obligations.
Funding
This study was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP22K05739 (to CY), and JP18H02246 and JP22H02391 (to KT).
Author information
Authors and Affiliations
Contributions
CY, CM, and KT conceptualized the research. CY, KT, and NM prepared the study materials and collected the data. CM, HI, and KE contributed to the planning of experimental strategies. CY and KT wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised due to a retrospective open access order.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oda-Yamamizo, C., Mitsuda, N., Milkowski, C. et al. Heterologous gene expression system for the production of hydrolyzable tannin intermediates in herbaceous model plants. J Plant Res 136, 891–905 (2023). https://doi.org/10.1007/s10265-023-01484-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-023-01484-2