Abstract
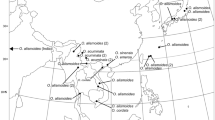
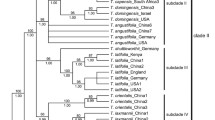
Sagittaria is a genus of ca. 40 species in the aquatic plant family Alismataceae with a nearly global distribution, and a center of diversity in the New World. Two thirds of the known species are native to the Americas, while only a few species are distributed in Africa, Asia and Europe. A previous biogeographic analysis of the genus suggested an African origin for the genus with subsequent dispersal to North America and then to East Asia. Here we expanded the taxon sampling with a focus on the New World taxa and applied species delimitation and biogeographic analyses to revise the knowledge of the phylogeny and evolution of the genus. We obtained largely similar topologies from the chloroplast DNA and nuclear DNA (ITS) data sets. The 74 accessions sampled for our analyses were delimited into 29 species and several cryptic taxa were revealed in widely distributed species. Biogeographic analysis supported basal diversification in South America and subsequent colonization to North America and Asia.




Similar content being viewed by others
References
Baldwin BG (1992) Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol Phylogenet Evol 1:3–16
Bogin C (1955) Revision of the genus Sagittaria (Alismataceae). Mem N Y Bot Gard 9:179–223
Bouckaert RR, Heled J, Kühnert D, Vaughan TG, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537
Chen LY, Chen JM, Gituru RW, Temam TD, Wang QF (2012) Generic phylogeny and historical biogeography of Alismataceae, inferred from multiple DNA sequences. Mol Phylogenet Evol 63:407–416
Crow GE, Hellquist CB (2000) Aquatic and wetland plants of northeastern North America. Two Monocotyledons: Angiosperms. University of Wisconsin Press, Madison
Cuenoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002) Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot 89:132–144
Dandy JE, Sagittaria L (1980) Flora Europaea. In: Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA, Chater AO, Richardson IBK (eds) Alismataceae to Orchidaceae, vol 5. Cambridge University Press, Cambridge, pp 1–2
de Candolle A (1855) Gèographie botanique raisonnèe; ou, exposition des faits principaux et des lois concernant la distribution gèographique des plantes de l’ èpoque actuelle, vol 2. Victor Masson, Paris, p 1365
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214–222
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Haynes RR, Hellquist CB (2000) Alismataceae. In: Flora of North America Editorial Committee (Eds.). Flora of North America North of Mexico. vol 22. Oxford University Press, Oxford, pp 7−25
Haynes RR, Holm-Nielsen LB (1994) The Alismataceae. Flora neotropica. Monogr 64:1–112
Haynes RR, Holm-Nielsen LB (1995) Alismataceae. In: Berry PE, Holst BK, Yatskievych K (eds) Flora of venezuelan guayana, vol 2. Missouri Botanical Garden Press, St. Louis, pp 377–383
Holm-Nielsen LB, Haynes RR (1986) 191. Alismataceae. In: Harling G, Andersson L (eds) Flora of ecuador, vol 26. Publishing House of the Swedish Research Councils, Stockholm, pp 1–24
Ito Y, Ohi-Toma T, Murata J, Tanaka N (2010) Hybridization and polyploidy of an aquatic plant, Ruppia (Ruppiaceae), inferred from plastid and nuclear DNA phylogenies. Am J Bot 97:1156–1167
Ito Y, Ohi-Toma T, Murata J, Tanaka N (2013) Comprehensive phylogenetic analyses of the Ruppia maritima complex focusing on taxa from the Mediterranean. J Plant Res 126:753–762
Ito Y, Tanaka N, Ohi-Toma T, Murata J, Muasya AM (2015) Phylogeny of Ruppia (Ruppiaceae) revisited: molecular and morphological evidence for a new species from Western Cape, South Africa. Syst Bot 40:942–949
Ito Y, Tanaka N, Barfod AS, Bogner J, Li J, Yano O, Gale SW (2019) Molecular phylogenetic species delimitation in the aquatic genus Ottelia (Hydrocharitaceae) reveals cryptic diversity within a widespread species. J Plant Res 132:335–344
Jones GL (2017) Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J Math Biol 74:447–467
Jones GL, Aydin Z, Oxelman B (2015) DISSECT: an assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics 31:991–998
Kaplan Z (2002) Phenotypic plasticity in Potamogeton (Potamogetonaceae). Folia Geobot 37:141–170
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Keener BR (2005) Molecular systematics and revision of the aquatic monocot genus Sagittaria (Alismataceae). Dissertation, University of Alabama
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics, btz305
Lehtonen S (2018) Alismataceae. In: Ramella L (ed) Flora del paraguay, vol 49. Conservatoire et Jardin botaniques de la Ville de Genève, Geneve, pp 1–42
Lehtonen S, Myllys L (2008) Cladistic analysis of Echinodorus (Alismataceae): simultaneous analysis of molecular and morphological data. Cladistics 24:218–239
Les DH, Crawford DJ, Kimball RT, Moody ML, Landolt E (2003) Biogeography of discontinuously distributed hydrophytes: a molecular appraisal of intercontinental disjunctions. Intl J Plant Sci 164:917–932
Li Z, Ngarega BK, Lehtonen S, Gichira AW, Karichu MJ, Wang QF, Chen JM (2020) Cryptic diversity within the African aquatic plant Ottelia ulvifolia (Hydrocharitaceae) revealed by population genetic and phylogenetic analyses. J Plant Res 133:373–381
Little DP, Barrington DS (2003) Major evolutionary events in the origin and diversification of the fern genus Polystichum (Dryopteridaceae). Am J Bot 90:508–514
Matzke NJ (2013) Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front Biogeogr 5:242–248
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), 14 Nov 2010, New Orleans, pp 1–8. http://www.phylo.org/sub_sections/portal/cite.php
Nylander JAA (2002) MrModeltest. Ver. 1.0. Program distributed by the author. Department of Systematic Zoology, Uppsala University, Uppsala. Retrieved from https://www.ebc.uu.se/systzoo/staf/nylander.html
Proctor GR (2005) Family 4. Alismataceae water-plantain family. In: Acevedo-Rodriguez P, Strong MT (eds) Monocotyledons and gymnosperms of Puerto Rico and the Virgin Islands. Contributions from the United States national Herbarium, vol 52. Department of Botany, National Museum of Natural History, Washington, pp 58–63
Rambaut A (2009) FigTree ver. 1.3.1: Tree Figure Drawing Tool. Retrieved from https://tree.bio.ed.ac.uk/software/fgtree/. Accessed 4 Jan 2011
Rambaut A, Suchard MA, Xie W, Drummond AJ (2014) Tracer. ver. 1.6. Retrieved from https://beast.bio.ed.ac.uk/Tracer. Accessed 8 Sept 2018
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efcient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Sculthorpe CD (1967) The biology of aquatic vascular plants. Edward Arnold, London
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihoodbased phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771
Swoford DL (2002) PAUP: Phylogenetic analysis using parsimony (and other methods). ver. 40b10. Sinauer Associates, Sunderland
Tanaka N (2015) Alismataceae. In: Ohashi H, Kadota Y, Murata J, Yonekura K, Kihara H (eds) Wild flowers of Japan, vol 1. Heibonsha, Tokyo, pp 115–117 ((in Japanese))
Wang QF, Guo YH, Haynes RR, Hellquist CB (2010) Alismataceae. In: Wu C-Y, Raven PH, Hong D-Y (eds) Flora of China, vol 23. Science Press. Beijing & Missouri Botanical Garden Press, St. Louis, pp 84–89
Whitten WM, Williams NH, Chase MW (2000) Subtribal and generic relationships of Maxillarieae (Orchidaceae) with emphasis on Stanhopeinae: combined molecular evidence. Am J Bot 87:1842–1856
Wolf PG, Soltis PS, Soltis DE (1994) Phylogenetic relationships of Dennstaedtioid ferns: evidence from rbcL sequences. Mol Phylogenet Evol 3:383–392
Yang Z, Rannala B (1997) Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol Biol Evol 14:717–724
Yu Y, Blair C, He XJ (2020) RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol Biol Evol 37:604–606
Acknowledgements
The authors thank Drs. P. G. Aceñolaza (Buenos Aires), M. Sasagawa (Yamagata), and O. Yano (Okayama) for assistance in the field; D. Boufford and E. Wood (GH) and J. V. Freudenstein (OS) for hospitality during the first author’s recent visits; and C. Ishii (Tsukuba) for help with DNA sequencing. This research was partly supported by the international cooperative project “Biological inventory with special attention to Myanmar: Investigations of the origin of southern elements of Japanese flora and fauna” as the integrated research initiated by the National Museum of Nature and Science, Japan based on MoU between Forest Department, Ministry of Natural Resources and Environmental Conservation, Myanmar to NT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ito, Y., Tanaka, N., Keener, B.R. et al. Phylogeny and biogeography of Sagittaria (Alismataceae) revisited: evidence for cryptic diversity and colonization out of South America. J Plant Res 133, 827–839 (2020). https://doi.org/10.1007/s10265-020-01229-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-020-01229-5