Abstract
Plants have endogenous biological clocks that allow organisms to anticipate and prepare for daily and seasonal environmental changes and increase their fitness in changing environments. The circadian clock in plants, as in animals and insects, mainly consists of multiple interlocking transcriptional/translational feedback loops. The circadian clock can be entrained by environmental cues such as light, temperature and nutrient status to synchronize internal biological rhythms with surrounding environments. Output pathways link the circadian oscillator to various physiological, developmental, and reproductive processes for adjusting the timing of these biological processes to an appropriate time of day or a suitable season. Recent genomic studies have demonstrated that polymorphism in circadian clock genes may contribute to local adaptations over a wide range of latitudes in many plant species. In the present review, we summarize the circadian regulation of biological processes throughout the life cycle of plants, and describe the contribution of the circadian clock to local adaptation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
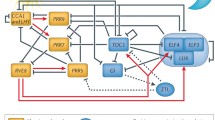
Plants as sessile organisms must precisely perceive environmental cues such as light and temperature to adapt their growth and development to surrounding environments. Circadian clocks are endogenous time-keeping mechanisms that allow organisms to anticipate and prepare for daily and seasonal changes in surrounding environments. Plants adjust the timing of various physiological, developmental, and reproductive processes to a proper time of day or an appropriate season based on the day length measured by the circadian clock (McClung 2006). Indeed, it has been reported that the circadian clock in plants achieves higher survival advantage and fitness (Dodd et al. 2005; Green et al. 2002). The circadian clock in plants mainly consists of interlocked transcriptional/translational feedback loops similar to the clocks in mammals or insects (Harmer et al. 2001). Previous studies have identified the core circadian clock genes, including CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), PSEUDO RESPONSE REGULATOR (PRR) family genes, TIMING OF CAB EXPRESSION 1 (TOC1), GIGANTEA (GI), EARLY FLOWERING (ELF) genes, and LUX ARRHYTHMO (LUX), which form multiple interlocked negative feedback loops (Nohales and Kay 2016). In addition, positive regulators such as REVEILLE (RVE) genes, LIGHT-REGULATED WD (LWD) genes, and NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED (LNK) genes have also been identified (Rawat et al. 2011; Rugnone et al. 2013; Wu et al. 2016).
The circadian clock in plants can be entrained by environmental cues, such as light, temperature, and nutrient status through multiple input pathways (Inoue et al. 2017). Then, the clock regulates various biological processes at an appropriate time of day through output pathways. Recent studies using natural accessions have revealed that altering circadian timing due to natural variation in circadian clock genes could contribute to the adaptation to local environments over a wide range of latitudes. In this review, we focus on output responses regulated by the circadian clock (Fig. 1). We summarize the circadian regulation of biological processes throughout the life cycle of plants at the cellular, tissue/organ, and individual levels.
The circadian clock in plants regulates various biological processes throughout the life cycle. The circadian clock adjusts the timing of output responses, such as petal opening, photosynthesis, stomatal opening, leaf movement, flowering, cell cycle progression, and hypocotyl elongation to an appropriate time of day
The role of the circadian clock at the cellular level processes
The circadian clock system is found not only in multicellular organisms but also in unicellular organisms. It has been reported that cell cycle of the cyanobacterium Synechococcus elongatus exhibits circadian gating, and therefore is probably regulated by the circadian clock (Mori et al. 1996; Yang et al. 2010). A time-lapse imaging revealed that phosphorylation state of the oscillator protein KaiC that is associated with elevated ATPase activity applies a circadian checkpoint on the cell division in S. elongatus by inhibiting FtsZ ring formation (Dong et al. 2010). Intriguingly, circadian rhythms are sustained in cells that divide three or more times during one circadian period, suggesting that cell division cycling does not interfere with the circadian clock of S. elongatus (Kondo et al. 1997). Clock-regulated cell cycle progression is also found in the unicellular red algae Cyanidioschyzon merolae. The cell cycle of C. merolae is highly synchronized with external light/dark cycle in which cell division occurred during the dark period (Imoto et al. 2011; Suzuki et al. 1994). The phosphorylation of the E2F protein, a key regulator for G1/S transition, is regulated by the circadian clock, resulting in the restriction of the G1/S transition to early subjective night. Furthermore, the uncoupling of cell cycle progression from circadian rhythms decreases the growth rate probably due to high oxidative stress, suggesting that the restriction of cell cycle progression during the night might be important to unicellular photosynthetic eukaryotes (Miyagishima et al. 2014). Given that the temporal circadian gating of cell division has been observed in a wide variety of eukaryotic lineages from unicellular organisms to mammals (Johnson 2010), the cell cycle of eukaryotes is closely associated with the circadian clock during the course of evolution. Although the circadian clock-coupled cell division is reported in many organisms, there is no solid evidence that the circadian clock in plants regulates cell cycle progression. However, a recent report demonstrated that the cell size of mesophyll cells in the leaf is altered in circadian clock mutants. The size of rve 4 6 8 triple and rve 3 4 5 6 8 quintuple mutants is increased compared with that of the wild-type plants at both seedling and adult stages of development. The average area of mesophyll cells in the leaves of rve4 6 8 triple mutants is approximately 1.3-fold larger than that in the wild-type plants, suggesting that the increased size in the rve mutants is primarily due to the larger average cell size (Gray et al. 2017). Given that cell size is closely associated with the length of cell cycle (Jones et al. 2017), it is possible that the circadian clock in plants could regulate cell cycle progression as well as that in other species.
Another circadian regulation of cellular level processes is observed in stomatal opening and photosynthesis. In Arabidopsis thaliana (Arabidopsis) and the bean Phaseolus vulgaris, stomatal conductance is higher during the day than at night (Hennessey et al. 1993; Somers et al. 1998), whereas the stomata in some crassulacean acid metabolism plants Hoya are more open at night than during the day (Thimann et al. 1992). Additionally, photosynthesis is also regulated by the circadian clock (Dodd et al. 2014b). The expression of a large set of genes involved in the light-harvesting complex is regulated by the circadian clock (Harmer et al. 2000; Schaffer et al. 2001). Consistent with the rhythmic expression of mRNA, the accumulation level of the chlorophyll a/b binding protein (CAB) and ribulose 1,5-bisphosphate carboxylase/oxygenase activase (RCA) proteins in the tomato Lycopersicon esculentum Mill is under the control of the circadian clock (Martinocatt and Ort 1992; Pilgrim and Mcclung 1993). Furthermore, phosphorylation of the D1 photosystem II reaction center protein in the duckweed Spirodela oligorrhiza and phosphoenolpyruvate carboxylase in crassulacean acid metabolism plants is also regulated by the circadian clock (Booij-James et al. 2002; Borland et al. 1999; Hartwell et al. 1999; Nimmo 1998, 2000), indicating that photosynthesis is strictly regulated by the circadian clock at the transcriptional, post-transcriptional, and post-translational levels. In addition to photosynthesis, photorespiration, sugar metabolism, and starch degradation are also regulated by the circadian clock (Harmer et al. 2000; Lu et al. 2005; McClung et al. 2000). It has been suggested that circadian control of photosynthesis and physiology provides higher fitness for plants (Dodd et al. 2005). However, recent reports demonstrate that the circadian control of photosynthesis is not sufficient to explain the effects of the circadian clock on plant fitness. Instead, circadian control of starch degradation during the night appears to be important (Dodd et al. 2014a; Graf et al. 2010; Graf and Smith 2011), although how the circadian clock controls the rate of starch degradation during the night remains unknown.
The role of the circadian clock at the tissue/organ level during plant development
The circadian clock in plants regulates various developmental processes throughout the life cycle of plants. At the earliest stage of plant development, the circadian clock regulates seed germination. The photoperiodic control of seed germination has been reported in many plant species (Baskin and Baskin 1976; Black and Wareing 1954, 1955; Densmore 1997), suggesting the existence of functional circadian system in seeds at least in some plant species. Indeed, imbibition, but not release from stratification can reset the circadian clock and synchronize the clocks among population of seedlings (Zhong et al. 1998). Furthermore, the circadian control of gas exchange is observed in free-running condition in dry onion seeds (Bryant 1972), suggesting that the functional circadian system is present even in quiescent seeds before germination. More recent reports demonstrated that the circadian clock controls seed germination probably through regulating a series of abscisic acid- and gibberellin-related genes expression (Covington et al. 2008; Penfield and Hall 2009).
The circadian clock in plants affects many aspects of plant growth. Hypocotyl elongation is a well-characterized growth regulated by the circadian clock. The circadian regulation of rhythmic elongation is observed in constant light (Dowson-Day and Millar 1999), although hypocotyl elongation is arrhythmic in constant dark (Nozue et al. 2007), suggesting that light signal is essential for the circadian regulation of hypocotyl elongation. This rhythmic growth is dependent on the function of two basic helix-loop-helix transcription factors, PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PIF5. PIF4 and PIF5 act as a signaling hub to integrate various external and internal cues, such as light, clock, temperature, phytohormone, and sucrose signaling (de Lucas et al. 2008; de Montaigu et al. 2010; Feng et al. 2008; Leivar and Monte 2014; Stewart et al. 2011). It has been demonstrated that expression levels of PIF4 and PIF5 are well correlated with hypocotyl growth rate (Nozue et al. 2007). ELF3, ELF4 and LUX tripartite complex represses the expression of PIF4 and PIF5 in the early evening (Nusinow et al. 2011). In addition, ELF3 physically interacts with PIF4 to inhibit its transcriptional activity in the early night (Nieto et al. 2015). Furthermore, PIF4 and PIF5 are rapidly degraded through the interaction with light-activated PHYTOCHROME B (PHYB) upon the irradiation of light (Lorrain et al. 2008). Thus, the activity of PIF4 and PIF5 is restricted to the late night, resulting in the photoperiodic regulation of hypocotyl elongation before dawn under diurnal conditions (Niwa et al. 2009). A similar circadian regulation of elongation has been reported in other plant species such as the tomato L. esculentum and the red goosefoot Chenopodium rubrum (Fernandez and Wagner 1994; Lecharny and Wagner 1984; Tukey and Ketellapper 1963). Another circadian regulation of growth is observed in diel leaf movements. In legumes including Mimosa pudica, leaf movements occurred by expansion and contraction of specialized cells at the base of the petiole called the pulvinus that allows for rapid reversible changes in leaf position (Uehlein and Kaldenhoff 2008; Whippo and Hangarter 2009). Plants without pulvini, however, also undergo circadian leaf movements that at least partially depend on antiphasic differential growth of the abaxial and adaxial sides of the leaf blades and petioles (Polko et al. 2012; Rauf et al. 2013; Uehlein and Kaldenhoff 2008). Such circadian regulation of leaf movements have been observed in several plant species, such as Arabidopsis, Brassica oleracea, B. rapa, tobacco, and potato (Engelmann et al. 1992; Lou et al. 2011; Salathia et al. 2007; Siefritz et al. 2004; Yanovsky et al. 2000). It has been recently reported that ELF3 is required to maintain the proper phase of leaf growth and movements, although PIF4 and PIF5 are not essential to sustain rhythmic leaf growth (Dornbusch et al. 2014). These findings suggest that molecular mechanisms underlying rhythmic hypocotyl and leaf growth are different from each other. In addition, the rate of circumnutations and shade avoidance response is also under the control of the circadian clock (Niinuma et al. 2005; Salter et al. 2003; Stolarz 2009; Takase et al. 2013; Whippo and Hangarter 2009). Recently, heliotropism, solar tracking movements, in the sunflower Helianthus annuus is driven by antiphasic patterns of elongation on the east and west sides of the stem regulated through the coordinate action of light-signaling pathways and the circadian clock (Atamian et al. 2016).
Photoperiodic flowering is the most characterized developmental event regulated by the circadian clock (Endo et al. 2016; Shim et al. 2017). CONSTANS (CO), a key transcription factor for photoperiodic flowering, is strictly regulated by the circadian clock and light signaling pathways (Putterill et al. 1995; Samach et al. 2000; Suarez-Lopez et al. 2001; Valverde et al. 2004). CYCLING DOF FACTORs (CDFs), whose expression pattern is under the control of the circadian clock repress CO transcription in the morning (Imaizumi et al. 2005). Only under long-day conditions, CDFs are degraded by a complex of GI and FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1). The peak times of GI and FKF expression differ under short-day conditions, whereas the peaks of both expressions coincide in late afternoon under long-day conditions. Furthermore, blue-light activates FKF1 and stabilizes the GI-FKF1 complex by enhancing the interaction of GI with FKF1 before dusk. These internal and external coincidence allows the GI-FKF1 complex to target CDFs for proteasomal degradation only in the late afternoon of long-days (Sawa et al. 2007), thereby derepressing CO (Song et al. 2012). The stability of CO protein is regulated through light signaling pathways mediated by multiple photoreceptors. CO protein is degraded by a complex of CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and SUPPRESSOR OF PHYA-105 1 (SPA1) during the dark (Jang et al. 2008; Laubinger et al. 2006). In addition, CO protein is destabilized by phyB through two distinct mechanisms mediated by HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1 (HOS1) and PHYTOCHROME-DEPENDENT LATE-FLOWERING (PHL) in the morning (Endo et al. 2013; Lazaro et al. 2015), whereas phyA and CRYPTOCHROME 2 (CRY2) stabilize CO protein in the late afternoon probably by inhibiting the function of the COP1-SPA complex (Sheerin et al. 2015; Zuo et al. 2011). This coordinated action of the circadian clock and light signaling pathways allows for accumulation of CO protein in the late afternoon under long-day conditions, and then CO induces the transcription of a florigen-encoding FLOWERING LOCUS T (FT) for photoperiodic flowering (Tiwari et al. 2010). Consistent with previous findings that phyA, CRY2, COP1, SPA1, and CO all function in the phloem companion cells for photoperiodic flowering (An et al. 2004; Endo et al. 2007; Jang et al. 2008; Kirchenbauer et al. 2016; Ranjan et al. 2011), recent reports demonstrated that the circadian clock in the phloem companion cells, not in the mesophyll, epidermis, and shoot apex cells, is critical for photoperiodic flowering, suggesting the significance of vasculature-specific clock function for photoperiodic flowering (Endo et al. 2014; Shimizu et al. 2015).
At the late stage of plant development after flowering, the circadian clock regulates flower opening for successful pollination. The circadian clock restricts the timing of petal opening to a part of the day when potential pollinators are most active. In Arabidopsis, flower opens in the morning and closes at midday (van Doorn and Kamdee 2014; van Doorn and van Meeteren 2003), whereas flower of night-blooming Cestrum nocturnum opens in the evening and closes at dawn (Overland 1960; van Doorn and Kamdee 2014). Furthermore, some volatile compounds and nectar for attraction of pollinators are also regulated by the circadian clock, and therefore they are emitted in the correct timing of potential pollinator activities during the day (Kolosova et al. 2001; Pesti 1976; Verdonk et al. 2003). The rhythms of these compounds are likely generated through the circadian regulation of their biosynthetic genes expression (Fenske et al. 2015; Fenske and Imaizumi 2016).
The role of the circadian clock for adaptation to local environments at the individual level
The circadian clock regulates various cellular or developmental processes and provides higher fitness under diurnal conditions as described above. Many plant species including Arabidopsis spread into different climatic and latitudinal areas with a wide range of day-length changes throughout the year. Several reports have suggested that mutations in the clock genes could contribute to adaptation to local environments. The period length of leaf movements in 150 Arabidopsis accessions is positively correlated with the day length at the latitude of origin, implying the adaptive significance of the circadian clock (Michael et al. 2003). Quantitative trait loci (QTL) analysis revealed that multiple loci interact to determine the period length, phase, and amplitude of leaf movements. PRR7 was located at some of the QTL for circadian period, and mutants defective in PRR family genes exhibit altered circadian period, suggesting that polymorphism in the PRR genes is a candidate for adaptation to local environments. Positive correlation between the period length of leaf movements and the day length at the latitude of origin is also reported in Mimulus guttatus and Glycine max (Greenham et al. 2017). Similarly, the period length of core clock gene expression in Capsella bursa-pastris ecotypes is strongly correlated with the latitudinal origin (Slotte et al. 2007). C. bursa-pastris ecotypes derived from lower latitudes showed earlier flowering, indicating that the phase advance in clock gene expression could contribute to early flowering. Polymorphism in ELF3 originated in Central Asia causes a short-period under light and severely dampened oscillation in the dark because of the defects in nuclear localization of ELF3, suggesting the contribution of mutations in ELF3 to local adaptation (Anwer et al. 2014).
Perspectives
The circadian clock regulates various cellular and developmental processes throughout the life cycle of plants, and indeed other organisms. Although accumulating evidence indicates the significance of the circadian clock for plant fitness, how the circadian system is able to regulate so many output processes and contribute to higher fitness is still largely unknown. However, recent advances of chromatin immunoprecipitation combined with massively parallel sequencing (ChIP-seq) achieved genome-wide identification of the direct regulations of clock-output genes by some of the core clock components (Ezer et al. 2017; Kamioka et al. 2016; Liu et al. 2016; Nagel et al. 2015; Nakamichi et al. 2012). Furthermore, recent analyses revealed that plants have decentralized oscillator networks consisting of multiple tissue-specific clocks, which show asymmetric couplings (Endo et al. 2014; Shimizu et al. 2015; Takahashi et al. 2015). Therefore, it is important to know how oscillator networks integrate spatial and temporal information into the whole body to regulate various responses.
As described above, polymorphism in the circadian clock genes may contribute to the adaptation to local environments. Indeed, many domesticated crops contain mutations in the core clock genes, resulting in the optimized flowering time (Nakamichi 2015). Since the day length for a given latitude is invariable in spite of the global climate changes, optimization of flowering time will continue to be needed for further crop domestication. Although modification of the circadian clock genes may enhance crop growth and in turn yields, circadian transcriptome data of crop species is still not readily available. Moreover, recent modeling of transcriptome data in field conditions versus in a growth chamber revealed that the progression of internal time in the morning and evening is different in the field and chamber probably due to the difference in increasing and decreasing rate of light intensity and temperature (Matsuzaki et al. 2015), suggesting the importance of integrating the parameters derived from controlled growth chamber into field conditions. Time-course sequences of transcriptome data of crop species in field conditions and mathematical modeling could help us to determine new targets for improving crop yields and provide new insights into the role of the circadian clock in fluctuating environments for local adaptation.
Change history
21 February 2018
The article” Circadian clock during plant development”
References
An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, Coupland G (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131:3615–3626
Anwer MU, Boikoglou E, Herrero E, Hallstein M, Davis AM, James GV, Nagy F, Davis SJ (2014) Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 3:e02206
Atamian HS, Creux NM, Brown EA, Garner AG, Blackman BK, Harmer SL (2016) Circadian regulation of sunflower heliotropism, floral orientation, and pollinator visits. Science 353:587–590
Baskin JM, Baskin CC (1976) Effect of photoperiod on germination of cyperus inflexus seeds. Bot Gaz 137:269–273
Black M, Wareing PF (1954) Photoperiodic control of germination in seed of Birch (Betula Pubescens Ehrh). Nature 174:705–706
Black M, Wareing PF (1955) Growth studies in woody species. 7. Photoperiodic control of germination in Betula Pubescens Ehrh. Physiol Plant 8:300–316
Booij-James IS, Swegle WM, Edelman M, Mattoo AK (2002) Phosphorylation of the D1 photosystem II reaction center protein is controlled by an endogenous circadian rhythm. Plant Physiol 130:2069–2075
Borland AM, Hartwell J, Jenkins GI, Wilkins MB, Nimmo HG (1999) Metabolite control overrides circadian regulation of phosphoenolpyruvate carboxylase kinase and CO2 fixation in Crassulacean acid metabolism. Plant Physiol 121:889–896
Bryant TR (1972) Gas-exchange in dry seeds—circadian rhythmicity in absence of DNA-replication, transcription, and translation. Science 178:634–636
Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9:R130
de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451:480–484
de Montaigu A, Toth R, Coupland G (2010) Plant development goes like clockwork. Trends Genet 26:296–306
Densmore RV (1997) Effect of day length on germination of seeds collected in Alaska. Am J Bot 84:274–278
Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309:630–633
Dodd AN, Dalchau N, Gardner MJ, Baek SJ, Webb AAR (2014a) The circadian clock has transient plasticity of period and is required for timing of nocturnal processes in Arabidopsis. New Phytol 201:168–179
Dodd AN, Kusakina J, Hall A, Gould PD, Hanaoka M (2014b) The circadian regulation of photosynthesis. Photosynth Res 119:181–190
Dong GG, Yang Q, Wang Q, Kim YI, Wood TL, Osteryoung KW, van Oudenaarden A, Golden SS (2010) Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140:529–539
Dornbusch T, Michaud O, Xenarios I, Fankhauser C (2014) Differentially phased leaf growth and movements in Arabidopsis depend on coordinated circadian and light regulation. Plant Cell 26:3911–3921
Dowson-Day MJ, Millar AJ (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17:63–71
Endo M, Mochizuki N, Suzuki T, Nagatani A (2007) CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19:84–93
Endo M, Tanigawa Y, Murakami T, Araki T, Nagatani A (2013) PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc Natl Acad Sci USA 110:18017–18022
Endo M, Shimizu H, Nohales MA, Araki T, Kay SA (2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515:419–422
Endo M, Araki T, Nagatani A (2016) Tissue-specific regulation of flowering by photoreceptors. Cell Mol Life Sci 73:829–839
Engelmann W, Simon K, Phen CJ (1992) Leaf movement rhythm in Arabidopsis thaliana. Z Naturforsch C 47:925–928
Ezer D, Jung JH, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai XL, Stockle D, Zubieta C, Jaeger KE, Wigge PA (2017) The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3:17087
Feng SH, Martinez C, Gusmaroli G, Wang Y, Zhou JL, Wang F, Chen LY, Yu L, Iglesias-Pedraz JM, Kircher S, Schafer E, Fu XD, Fan LM, Deng XW (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451:475–479
Fenske MP, Imaizumi T (2016) Circadian rhythms in floral scent emission. Front Plant Sci 7:462
Fenske MP, Hazelton KDH, Hempton AK, Shim JS, Yamamoto BM, Riffell JA, Imaizumi T (2015) Circadian clock gene LATE ELONGATED HYPOCOTYL directly regulates the timing of floral scent emission in Petunia. Proc Natl Acad Sci USA 112:9775–9780
Fernandez SR, Wagner E (1994) A new method of measurement and analysis of the stem extension growth rate to demonstrate complete synchronization of Chenopodium rubrum plants by environmental conditions. J Plant Physiol 144:362–369
Graf A, Smith AM (2011) Starch and the clock: the dark side of plant productivity. Trends Plant Sci 16:169–175
Graf A, Schlereth A, Stitt M, Smith AM (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107:9458–9463
Gray JA, Shalit-Kaneh A, Chu DN, Hsu PY, Harmer SL (2017) The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol 173:2308–2322
Green RM, Tingay S, Wang ZY, Tobin EM (2002) Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129:576–584
Greenham K, Lou P, Puzey JR, Kumar G, Arnevik C, Farid H, Willis JH, McClung CR (2017) Geographic variation of plant circadian clock function in natural and agricultural settings. J Biol Rhythms 32:26–34
Harmer SL, Hogenesch LB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290:2110–2113
Harmer SL, Panda S, Kay SA (2001) Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol 17:215–253
Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GL, Nimmo HG (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20:333–342
Hennessey TL, Freeden AL, Field CB (1993) Environmental effects of circadian rhythms in photosynthesis and stomatal opening. Planta 189:369–376
Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1F-BOX protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293–297
Imoto Y, Yoshida Y, Yagisawa F, Kuroiwa H, Kuroiwa T (2011) The cell cycle, including the mitotic cycle and organelle division cycles, as revealed by cytological observations. J Electron Microsc 60:S117–S136
Inoue K, Araki T, Endo M (2017) Integration of input signals into the gene network in the plant circadian clock. Plant Cell Physiol 58:977–982
Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27:1277–1288
Johnson CH (2010) Circadian clocks and cell division What’s the pacemaker? Cell Cycle 9:3864–3873
Jones AR, Forero-Vargas M, Withers SP, Smith RS, Traas J, Dewitte W, Murray JAH (2017) Cell-size dependent progression of the cell cycle creates homeostasis and flexibility of plant cell size. Nat Commun 8:15060
Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28:696–711
Kirchenbauer D, Viczian A, Adam E, Hegedus Z, Klose C, Leppert M, Hiltbrunner A, Kircher S, Schafer E, Nagy F (2016) Characterization of photomorphogenic responses and signaling cascades controlled by phytochrome—a expressed in different tissues. New Phytol 211:584–598
Kolosova N, Gorenstein N, Kish CM, Dudareva N (2001) Regulation of circadian methyl benzoate emission in diurnally and nocturnally emitting plants. Plant Cell 13:2333–2347
Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS (1997) Circadian rhythms in rapidly dividing cyanobacteria. Science 275:224–227
Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133:3213–3222
Lazaro A, Mouriz A, Pineiro M, Jarillo JA (2015) Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27:2437–2454
Lecharny A, Wagner E (1984) Stem extension rate in light-grown plants—evidence for an endogenous circadian rhythm in Chenopodium rubrum. Physiol Plant 60:437–443
Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26:56–78
Liu TL, Newton L, Liu MJ, Shiu SH, Farre EM (2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170:528–539
Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53:312–323
Lou P, Xie Q, Xu X, Edwards CE, Brock MT, Weinig C, McClung CR (2011) Genetic architecture of the circadian clock and flowering time in Brassica rapa. Theor Appl Genet 123:397–409
Lu Y, Gehan JP, Sharkey TD (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol 138:2280–2291
Martinocatt S, Ort DR (1992) Low-temperature interrupts circadian regulation of transcriptional activity in chilling-sensitive plants. Proc Natl Acad Sci USA 89:3731–3735
Matsuzaki J, Kawahara Y, Izawa T (2015) Punctual transcriptional regulation by the rice circadian clock under fluctuating field conditions. Plant Cell 27:633–648
McClung CR (2006) Plant circadian rhythms. Plant Cell 18:792–803
McClung CR, Hsu M, Painter JE, Gagne JM, Karlsberg SD, Salome PA (2000) Integrated temporal regulation of the photorespiratory pathway. Circadian regulation of two arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiol 123:381–391
Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302:1049–1053
Miyagishima SY, Fujiwara T, Sumiya N, Hirooka S, Nakano A, Kabeya Y, Nakamura M (2014) Translation-independent circadian control of the cell cycle in a unicellular photosynthetic eukaryote. Nat Commun 5:3807
Mori T, Binder B, Johnson CH (1996) Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 h. Proc Natl Acad Sci USA 93:10183–10188
Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112:E4802–E4810
Nakamichi N (2015) Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol 56:594–604
Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109:17123–17128
Nieto C, Lopez-Salmeron V, Daviere JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol 25:187–193
Niinuma K, Someya N, Kimura M, Yamaguchi I, Hamamoto H (2005) Circadian rhythm of circumnutation in inflorescence stems of Arabidopsis. Plant Cell Physiol 46:1423–1427
Nimmo HG (1998) Circadian regulation of a plant protein kinase. Chronobiol Int 15:109–118
Nimmo HG (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5:75–80
Niwa Y, Yamashino T, Mizuno T (2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol 50:838–854
Nohales MA, Kay SA (2016) Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23:1061–1069
Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448:358–361
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402
Overland L (1960) Endogenous rhythm in opening and odor of flowers of Cestrum nocturnum. Am J Bot 47:378–382
Penfield S, Hall A (2009) A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21:1722–1732
Pesti J (1976) Daily fluctuations in sugar content of nectar and periodicity of secretion in Compositae. Acta Agron Hung 25:5–17
Pilgrim ML, Mcclung CR (1993) Differential involvement of the circadian clock in the expression of genes required for ribulose-1,5-bisphosphate carboxylase oxygenase synthesis, assembly, and activation in Arabidopsis thaliana. Plant Physiol 103:553–564
Polko JK, van Zanten M, van Rooij JA, Maree AFM, Voesenek LACJ, Peeters AJM, Pierik R (2012) Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol 193:339–348
Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc-finger transcription factors. Cell 80:847–857
Ranjan A, Fiene G, Fackendahl P, Hoecker U (2011) The Arabidopsis repressor of light signaling SPA1 acts in the phloem to regulate seedling de-etiolation, leaf expansion and flowering time. Development 138:1851–1862
Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B (2013) NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 25:4941–4955
Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7:e1001350
Rugnone ML, Faigon Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Mas P, Yanovsky MJ (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110:12120–12125
Salathia N, Lynn JR, Millar AJ, King GJ (2007) Detection and resolution of genetic loci affecting circadian period in Brassica oleracea. Theor Appl Genet 114:683–692
Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426:680–683
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318:261–265
Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13:113–123
Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27:189–201
Shim JS, Kubota A, Imaizumi T (2017) Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173:5–15
Shimizu H, Katayama K, Koto T, Torii K, Araki T, Endo M (2015) Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nat Plants 1:15163
Siefritz F, Otto B, Bienert GP, van der Krol A, Kaldenhoff R (2004) The plasma membrane aquaporin NtAQP1 is a key component of the leaf unfolding mechanism in tobacco. Plant J 37:147–155
Slotte T, Holm K, McIntyre LM, Lagercrantz U, Lascoux M (2007) Differential expression of genes important for adaptation in Capsella bursa-pastoris (Brassicaceae). Plant Physiol 145:160–173
Somers DE, Webb AAR, Pearson M, Kay SA (1998) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125:485–494
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336:1045–1049
Stewart JL, Maloof JN, Nemhauser JL (2011) PIF genes mediate the effect of sucrose on seedling growth dynamics. PLoS One 6:e19894
Stolarz M (2009) Circumnutation as a visible plant action and reaction: physiological, cellular and molecular basis for circumnutations. Plant Signal Behav 4:380–387
Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120
Suzuki K, Ehara T, Osafune T, Kuroiwa H, Kawano S, Kuroiwa T (1994) Behavior of mitochondria, chloroplasts and their nuclei during the mitotic-cycle in the ultramicroalga Cyanidioschyzon merolae. Eur J Cell Biol 63:280–288
Takahashi N, Hirata Y, Aihara K, Mas P (2015) A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell 163:148–159
Takase M, Mizoguchi T, Kozuka T, Tsukaya H (2013) The unique function of the Arabidopsis circadian clock gene PRR5 in the regulation of shade avoidance response. Plant Signal Behav 8:e23534
Thimann KV, Tan ZY, Park J (1992) Cycling of stomatal aperture in leaves of plants with crassulacean acid metabolism under constant conditions. Am J Bot 79:23–27
Tiwari SB, Shen Y, Chang HC, Hou YL, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, Belachew A, Repetti PP, Reuber TL, Ratcliffe OJ (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187:57–66
Tukey HB, Ketellapper HJ (1963) Length of light-dark cycle and plant growth. Am J Bot 50:110–115
Uehlein N, Kaldenhoff R (2008) Aquaporins and plant leaf movements. Ann Bot 101:1–4
Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–1006
van Doorn WG, Kamdee C (2014) Flower opening and closure: an update. J Exp Bot 65:5749–5757
van Doorn WG, van Meeteren U (2003) Flower opening and closure: a review. J Exp Bot 54:1801–1812
Verdonk JC, de Vos CHR, Verhoeven HA, Haring MA, van Tunen AJ, Schuurink RC (2003) Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62:997–1008
Whippo CW, Hangarter RP (2009) The “sensational” power of movement in plants: a Darwinian system for studying the evolution of behavior. Am J Bot 96:2115–2127
Wu JF, Tsai HL, Joanito I, Wu YC, Chang CW, Li YH, Wang Y, Hong JC, Chu JW, Hsu CP, Wu SH (2016) LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat Commun 7:13181
Yang Q, Pando BF, Dong GG, Golden SS, van Oudenaarden A (2010) Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science 327:1522–1526
Yanovsky MJ, Izaguirre M, Wagmaister JA, Gatz C, Jackson SD, Thomas B, Casal JJ (2000) Phytochrome A resets the circadian clock and delays tuber formation under long days in potato. Plant J 23:223–232
Zhong HH, Painter JE, Salome PA, Straume M, McClung CR (1998) Imbibition, but not release from stratification, sets the circadian clock in Arabidopsis seedlings. Plant Cell 10:2005–2017
Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21:841–847
Acknowledgements
We thank T. Koto for technical assistance. This work was supported by JST PRESTO grant 888067 (to M.E.), JSPS KAKENHI grant 16H01240 and 17K19392 (to M.E.), grants from the Yamada Science Foundation, Senri Life Science Foundation, and the Nakajima Foundation (to M.E.), and Grants-in-Aid for Scientific Research on Priority Area 25113005 (to T.A.).
Author information
Authors and Affiliations
Corresponding author
Additional information
The original version of this article was revised due to a retrospective open access order.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Inoue, K., Araki, T. & Endo, M. Circadian clock during plant development. J Plant Res 131, 59–66 (2018). https://doi.org/10.1007/s10265-017-0991-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-017-0991-8