Abstract
Peritoneal mesothelioma (PeM) is an aggressive tumor with limited treatment options. The current study aimed to evaluate the value of next generation sequencing (NGS) of PeM samples in current practice. Foundation Medicine F1CDx NGS was performed on 20 tumor samples. This platform assesses 360 commonly somatically mutated genes in solid tumors and provides a genomic signature. Based on the detected mutations, potentially effective targeted therapies were identified. NGS was successful in 19 cases. Tumor mutational burden (TMB) was low in 10 cases, and 11 cases were microsatellite stable. In the other cases, TMB and microsatellite status could not be determined. BRCA1 associated protein 1 (BAP1) mutations were found in 32% of cases, cyclin dependent kinase inhibitor 2A/B (CDKN2A/B) and neurofibromin 2 (NF2) mutations in 16%, and ataxia-telangiectasia mutated serine/threonine kinase (ATM) in 11%. Based on mutations in the latter two genes, potential targeted therapies are available for approximately a quarter of cases (i.e., protein kinase inhibitors for three NF2 mutated tumors, and polyADP-ribose polymerase inhibitors for two ATM mutated tumors). Extensive NGS analysis of PeM samples resulted in the identification of potentially effective targeted therapies for about one in four patients. Although these therapies are currently not available for patients with PeM, ongoing developments might result in new treatment options in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peritoneal mesothelioma (PeM) is an aggressive tumor, arising from the peritoneum [1]. It comprises about ten to fifteen percent of all mesotheliomas, thereby being the second most common variant after pleural mesothelioma [2]. Due to its rarity and non-specific symptoms, it is often diagnosed at an advanced stage. Currently the best available treatment is a combination of cytoreductive surgery (CRS) with hyperthermic intraperitoneal chemotherapy (HIPEC) [3]. Unfortunately, most patients experience disease recurrence, even after complete cytoreduction. Adding (neo)adjuvant systemic chemotherapy to the treatment does not result in improved disease-free, or overall, survival [4], and only a small proportion of patients are eligible to undergo surgical treatment, while there is a lack of effective systemic treatment options [5].
Because PeM is so rare, it is especially hard to gather (randomized) evidence on the effect of new therapeutics. The heterogeneity of the tumor further complicates this research. Personalized strategies, based on tumor molecular characteristics, could be promising [6]. One approach is to identify potentially targetable mutations, which can be treated with readily available therapies. However, data on the mutational landscape of PeM have long been lacking. Recently, several studies have been published that provide more insights in the mutational profile of PeM [7,8,9,10,11]. These data could aid to identify new treatment options for patients with PeM. Preferably, these treatments are already registered for the treatment of (other) cancers, but currently there are also clinical trials that include patients based on tumor molecular characteristics rather than cancer type or location [12,13,14].
Foundation Medicine (FMI) offers a platform (Foundation One® CDx (F1CDx)) for next generation sequencing (NGS) of formalin fixed paraffin embedded (FFPE) tumor samples, which are often the only material available from diagnostic biopsies. The platform assesses a total of 360 genes that are known to be somatically mutated in solid tumors [15]. It also provides a genomic signature, by assessing tumor mutational burden (TMB) and microsatellite (in)stability (MSS/MSI). To evaluate the value of genomic characterization in patients with PeM in current daily practice, we performed broad targeted NGS on tumor biopsies from 20 patients who were referred to the Erasmus MC Cancer Institute from 2018 to 2021.
Methods
Patient selection and data handling
From 2018 to 2021, 41 PeM patients were referred to the Erasmus MC Cancer Institute in Rotterdam, a Dutch mesothelioma expert center. From these 41 patients, we identified 23 patients for whom excess tumor tissue was available and who provided permission to use this tissue for research purposes. NGS by Foundation Medicine (FMI) F1CDx was available for 20 tumor samples. To maximize the chance of finding new significant mutations, we further selected the patients based on sex, age and lack of asbestos exposure, thus enriching the cohort for females and younger patients [16]. All data were collected and managed according to the latest European privacy regulations (General Data Protection Regulation (GDPR), EU 2016/679). The study was approved by the EMC local ethics committee (MEC 2018-1286).
The foundation one® CDx assay
F1CDx uses DNA, acquired from FFPE tissue samples, for NGS of solid tumors. A comprehensive method description can be found in the technical information [15]. The assay is able to detect alterations in a total of 324 different genes, and another 36 introns of genes that are involved in rearrangements. Mutations in these genes and genetic rearrangements are known to occur in solid tumors and might be drive alterations for oncogenesis. Moreover, many of these mutations are susceptible to targeted therapies. A full list of included genes/rearrangements is rendered in the supplementary data (supplementary Table 1). The assay also determines the genetic signature of the tumor, by providing microsatellite status (MSI), and tumor mutational burden (TMB). MSI status is determined by genome wide analysis of 95 microsatellite loci. The assay report that is provided by Foundation One® also includes suggested (targeted) therapies or clinical trials for individual patients, based on latest available clinical evidence and an up-to-date overview of current clinical trials that include patients based on certain mutations.
Results
Patient and tumor characteristics
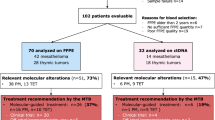
Broad targeted NGS on tumor biopsies from 20 individual patients was performed. Unfortunately, this resulted in one sample failure, leaving 19 samples to be fully analyzed. Table 1 provides a comprehensive overview of patient and disease characteristics per patient. The patients included in the study had a median age of 54 years (IQR 48–63), and three (15%) were female. Epithelioid morphology was most common, observed in 18 patients (90%), while sarcomatoid and biphasic morphology were each present in one patient (5%), as determined by an experienced subspecialist pathologist (JT) by histological analysis of hematoxylin/eosin (H&E) stained sections of FFPE tissue. A minority of patients (40%) had been (occupationally) exposed to asbestos in the past. The median peritoneal cancer index (PCI), a measure used to determine the extent of peritoneal disease, was 39 (IQR 31–39) [17]. Most patients (80%) presented with ascites at time of diagnosis and two patients (10%) had nodal dissemination. The Ki67 (or MIB) index reflects the percentage of proliferating cells and is a known prognostic indicator for PeM patients. Median Ki67 index was 8% (IQR 5–19%); while 11 tumors (58%) had a Ki67 index below 10% and eight tumors (42%) had a Ki67 index equal to or greater than 10%. Germline mutation analysis was performed in five out of 20 patients, of whom two patients were carrier of a BRCA associated protein 1 (BAP1) germline mutation.
Genomic signature
NGS data were available for 19 samples, as there was one sample failure (Table 1). The TMB could not be determined in nine (47%) cases due to low tumor purity. In all of the remaining cases (n = 10), TMB was low (defined as < 10 mutations/Mb). Similar outcomes were observed for MSI, which could not be determined in eight (42%) cases, and the remaining 11 tumors were microsatellite stable (MSS). In one patient, with a MSS tumor according to NGS, a frameshift mutation was detected in mutS homolog 6 (MSH6), encoding for the mismatch repair protein MSH6. Additional IHC for MMR proteins was performed on this sample, showing MLH-1, MSH-2, and PMS-2 proficiency and loss of MSH6 (supplementary Fig. 1). No germline analysis was performed for this patient. The most commonly affected gene in this cohort was BAP1, with oncogenic mutations found in six out of 19 patients (32%). In two samples, a variant of unknown significance (VUS) was detected in BAP1. Both cyclin dependent kinase inhibitor 2A/B (CDKN2A/B) and neurofibromin 2 (NF2) harbored mutations in three (16%) tumors. Genes harboring oncogenic mutations in this cohort are depicted in Fig. 1. Besides BAP1, CDKN2A/B, and NF2: ataxia-telangiectasia mutated serine/threonine kinase (ATM), polybromo 1 (PBRM1), protein kinase C iota (PRKCI), telomerase reverse transcriptase (TERT), and tumor protein p53 (TP53) were aberrant in ≥ 10% of the sequenced tumors. In Table 2, an overview of all affected genes is provided, including both significant mutations and VUS.
Variants of unknown significance
Besides known mutations involved in oncogenesis, the F1CDx analysis also provides a report of all VUSes. Variants in polymerase epsilon catalytic subunit (POLE), ROS proto-oncogene 1 receptor tyrosine kinase (ROS1), and zinc finger protein 703 (ZNF703) were determined to be a VUS in 15% of cases each. VUSes that were prevalent in ≥ 10% of cases were also included in Fig. 1. In two samples, a VUS in BAP1 was detected, resulting in loss of BAP1 expression at IHC.
Therapy recommendations
The analyses resulted in possible therapy recommendations for five patients (26%). All these recommendations were based on targeted therapies that were approved in the European Union for the treatment of other tumor types. None of these therapies is currently registered as a treatment for mesothelioma. For three (16%) patients with mutations in NF2, protein kinase inhibitor (PKI) therapy with either everolimus or temsirolimus could be of interest. For two (11%) other patients, therapy with polyADP-ribose polymerase (PARP)-inhibitors might be effective, based on mutations of the ATM gene.
Clinical trials
For patients with mutations in genes for which currently no targeted therapy is available, participation in clinical trials might be beneficial. Based on the NGS data, ten (53%) cases were possibly eligible to participate in clinical trials, based on thirteen observed mutations. Tumors with inactivating mutations, or loss of BAP1, are possibly susceptible to treatment with enhancer of zeste homolog 2 (EZH2) inhibitors. This resulted in a clinical trial recommendation for six (30%) cases with such a mutation. Two (11%) patients with mutations in ATM were possibly eligible to participate in various phase 1 and 2 clinical trials investigating ATR serine/threonine kinase (ATR) inhibitors, PARP inhibitors and/or DNA-dependent protein kinase catalytic subunit (DNA-PKcs) inhibitors. Another two (11%) patients were possibly eligible for participation in various clinical trials targeting focal adhesion kinase (FAK), programmed cell death 1 (PD1) and mammalian target of rapamycin complex 1/2 (mTORC1/C2) based on mutations in NF2. Mutations in phosphatase and tensin homolog (PTEN) and BRCA1 associated ring domain 1 (BARD1) resulted in similar recommendations, involving among others PARP and immune checkpoint inhibition. It should be noted that none of the patients in the current cohort participated in any of these trials, as these trials were not conducted in The Netherlands.
Discussion
The lack of effective treatments for peritoneal mesothelioma (PeM) makes it interesting to explore the use of targeted therapies that might benefit these patients. Although also rare, pleural mesothelioma is relatively more common and treatment strategies for PeM are commonly derived from the pleural variant. Recently, large cohorts of both pleural and PeM have provided more insights in their mutational profiles and provided possible targets or therapies [7,8,9,10,11, 18]. The mutational profile of the current study cohort is comparable to the TCGA pleural mesothelioma cohort, which is in line with the large cohorts of Hiltbrunner et al. and Dagogo-Jack et al [10, 11, 19].
To evaluate the value of broad NGS in patients with PeM in current practice, we performed broad targeted NGS on tumor biopsies from 20 individual PeM patients. Based on the molecular signature of these tumors, for about one in four patients, potentially effective targeted therapies are available. It should be noted that these targeted treatments have so far not been proven effective against mesothelioma (pleural or peritoneal). Therefore, the value of NGS in the current practice for these patients seems limited.
We did identify some clinical trials in which patients with PeM could potentially participate. There are also numerous ongoing trials in other tumor types that are investigating targeted therapies that might be beneficial for patients in our cohort based on the detected aberrations. As new targeted treatments, as well as combination therapies, are being continuously investigated, molecular characterization of individual patient tumors will be increasingly relevant in the future. Below, we reviewed biomarkers generated by NGS that could predict response to certain treatments and the most frequently mutated genes (i.e., oncogenic mutations in ≥ 10% of cases) in the current cohort, for which targeted therapies are currently available.
TMB and MSI status
TMB was low, and tumors were MSS in all cases for which this could be determined. For one patient in our cohort a MSH6 deficiency was reported. As MSI is a result of a deficient DNA MMR system, MSH6 deficient tumors are per definition MSI. Nonetheless, this tumor was reported as MSS by molecular MSI analysis. Several studies have indicated that molecular MSI analysis has lower sensitivity for MMR deficiency (dMMR) detection compared to IHC, which might be dependent on the origin of the primary tumor; hence, the value of molecular MSI analysis to detect dMMR tumors remains a subject of debate [20, 21]. Likewise, molecular MSI, but also TMB analysis, requires samples with sufficient tumor purity. Low tumor purity is an important challenge to these analyses in daily practice. Panel-based TMB estimation by targeted NGS has been proposed to result in a better estimate of the TMB, compared to the general method of measuring the TMB with the whole exome [22]. Moreover, increasing tumor purity by microdissection is valuable, but unfortunately not possible for send-out FMI tests.
Though MSI and TMB status could not be determined for eight and nine cases, respectively, it is likely that TMB and MSI are mostly low or absent in PeM. Arulananda and colleagues could not identify a single patient with MSI in a cohort of 335 patients with pleural mesothelioma, performed by IHC [23]. There are some studies that reported MSI in patients with mesothelioma, but these cases are rare [10, 24]. With regard to TMB, several studies reported low TMB in the majority of mesothelioma cases (both pleural and peritoneal) [10, 11, 25]. As both MSI and high-TMB tumors are associated with a good response to immune checkpoint inhibition (CPI) therapy, one might expect that these therapies are ineffective against mesothelioma [26]. Indeed, the recent checkmate 743 study by Baas et. al showed only modest responses to combination CPI therapy with nivolumab (anti-PD1) and ipilimumab (anti-CTLA4) as a first line treatment for pleural mesothelioma, although long term responders were established [27]. Hence, it is questionable whether MSI and TMB are optimal biomarkers to predict response to CPI.
Frequently aberrant genes
BAP1
BAP1 is the most frequently mutated gene found in mesothelioma (pleural and peritoneal), with about 30–50% of cases harboring somatic mutations. (AACR GENIE and COSMIC, February 2022) [28, 29]. Also, a significant proportion of PeM patients might be affected by the so-called ‘BAP1 tumor predisposition syndrome’ (BAP1-TPDS), as they are carriers of a germline BAP1 mutation [30]. Besides a predisposition for mesothelioma, these patients are also commonly affected by BAP1-inactivated melanocytic tumors, uveal melanoma, cutaneous melanoma and renal cell carcinoma [31]. In line with other studies, we found oncogenic BAP1 mutations in 32% of tumors in the current cohort, of which two patients were known carriers of a BAP1 germline mutation [7, 10, 11]. BAP1 encodes for the tumor suppressor protein ‘ubiquitin carboxyl-terminal hydrolase,’ which plays a role in several cellular processes involved in oncogenesis [32]. Though there are currently no treatments directly targeting BAP1, there are therapies targeting molecular pathways in which BAP1 is involved. BAP1 is associated with BRCA1 activation, thereby playing a key role in homologous recombination repair (HRR) [32,33,34]. Similar to ATM deficient tumors, BAP1 and BRCA1 deficient tumors might be susceptible to PARP inhibition and promising results have been reported in a phase 2 clinical trial [35]. However, in vitro results of sensitivity to PARP inhibition and its relationship to BAP1 status are inconsistent [36,37,38]. Another potential target is EZH2, which is upregulated in BAP1 deficient tumors. A preclinical showed increased sensitivity to EZH2 inhibition in BAP1 deficient mice [39]. A phase 2 trial including 74 patients with BAP1 deficient mesothelioma treated patients with PeM with the EZH2 inhibitor tazemetostat as a monotherapy [40]. A disease control rate of 51% at twelve weeks and 25% at 24 weeks was reported, but no complete and only two partial responses were observed. These modest responses do not seem to be related to BAP1 deficiencies and biomarkers to predict the response to tazemetostat have not yet been identified. Due to its involvement in HRR, BAP1 has also been studied as a biomarker for response to chemotherapy. Wildtype BAP1 has been associated with sensitivity to gemcitabine treatment in mesothelioma cell lines, but this has not been validated in patients with PeM [41, 42].
NF2
Based on several mutations in NF2, protein kinase inhibitors everolimus and temsirolimus could be a potential treatment option for 16% of patients in our cohort. NF2 is a tumor suppressor gene that plays an important role in cell proliferation and survival [43, 44]. NF2 is involved in the mammalian target of rapamycin (mTOR) signaling pathway. Inactivating mutations of NF2 lead to cell cycle progression and cell proliferation [45, 46]. NF2 mutations are reported by previous studies in around 25% of cases of PeM [10, 11]. Some clinical studies and some preclinical evidence suggest that NF2 inactivation might be associated with response to mTOR inhibitors.[47, 48] Everolimus and temsirolimus are both mTOR inhibitors and have been approved by the FDA for the treatment of neuroendocrine tumors of the gastro-intestinal tract or lung, HER2/neu-negative breast cancer and renal cell carcinoma, among others. A phase 2 study in pleural mesothelioma only showed a 2% response rate to everolimus [49]. This study, however, did not stratify patients based on mutational status. Considering that only about 15% of mesothelioma cases show mutations in NF2, the response rate might be higher when only these patients are included. However, some studies suggest that combination treatment might be indicated [50, 51].
ATM
Mutations in ATM were present in two patients in our cohort (11%), but were reported in only 2% of the patients in the large cohort of Hiltbrunner et al. [11]. Although rare, patients with PeM and mutations in ATM could benefit from treatment with PARP inhibitors. ATM is located on chromosome 11 and codes for the ATM serine/threonine kinase protein. This protein plays a role in the HRR pathway, among others by p53 activation, which has an important role in cell cycle arrest and apoptosis [52]. Mateo et al. found that deleterious ATM mutations in metastatic prostate cancer were associated with good response to olaparib, a PARP inhibitor that is approved for the treatment of several solid tumors in the European Union [35, 53]. However, the same group found no survival benefit for castration resistant prostate cancer patients, but these findings were the result of an underpowered interim analysis [54]. For other malignancies, such as gastric-cancer and renal cell carcinoma, similar relations between ATM mutations and response to PARP inhibition have been reported [55, 56]. Fennell et al. performed a phase 2 trial, treating 26 mesothelioma patients (25 pleural, 1 peritoneal) with the PARP inhibitor rucaparib after at least one cycle of systemic chemotherapy. They found a disease control rate of 58% at twelve weeks and 23% at 24 weeks, while toxicity was limited [57]. They selected patients with BAP1 and/or BRCA1 deficient tumors, other key proteins in HRR. HRR deficient tumors, such as ATM inactivated tumors, might have similar responses to PARP inhibition.
Strengths and limitations
The main strength of this study is the in-depth analysis of PeM molecular characteristics and the evaluation of its value in current daily practice. The current study provides more comprehensive data compared with recently published studies reporting on larger cohorts, which can be valuable for the guidance of future treatment strategies.[10, 11] Though our cohort only included 20 patients, with successful NGS in 19, PeM is such a rare tumor that data of its molecular characteristics remains valuable.
There are some limitations to the current study. As NGS was available for only 20 samples, we selected those patients that were most likely to harbor relevant mutations, resulting in selection bias. In addition, NGS requires sufficient amount of high-quality DNA. For NGS, FMI does not perform any tumor purification, requiring high-quality samples and resulting in a lower sensitivity for the detection of mutations. Selection of high-quality samples might also have resulted in selection bias. Despite this selection, there was one sample failure and TMB/MSI could not be determined in approximately half of the patients due to low tumor purity. This underlines the challenges of NGS in current daily practice, as the success of NGS highly depends on the sample quality and quantity. Despite low tumor purity, we were able to detect relevant mutations in the majority of patients. As the value of TMB/MSI in the treatment of patients with PeM seems limited, low tumor purity might not pose a serious problem in this patient population. Though not a limitation of the current study, another important factor to take into consideration with the interpretation of NGS data is tumor heterogeneity. Tumor heterogeneity results in the possibility of an unrepresentative tumor biopsy, which can be especially relevant in guiding possible treatment choices. Likewise, NGS often identifies variants of unknown significance (VUS), which have no clear clinical implications (yet). For example, one patient in our cohort [8] had a VUS in BAP1, but also showed loss of BAP1 on IHC, making it likely that this is actually a pathogenic mutation. Ongoing research will probably identify the nature of these mutations in the future.
Conclusion
The value of genomic characterization of PeM tumor samples in daily practice in the Netherlands is currently limited. NGS poses several practical challenges, and effective targeted therapies are limited. For about one in four patients in our cohort, NGS resulted in the identification of potentially effective targeted therapies that are currently available for other tumor types than PeM. Ongoing developments in targeted therapies will result in new treatment options, making genomic characterization increasingly relevant in the future.
Availability of data and materials
The data that support the findings of this study are included in the manuscript and any additional data are available from the corresponding author upon reasonable request.
References
Kim J, Bhagwandin S, Labow DM. Malignant peritoneal mesothelioma: a review. Ann Transl Med. 2017;5(11):236.
Boffetta P. Epidemiology of peritoneal mesothelioma: a review. Ann Oncol. 2007;18(6):985–90.
Yan TD, Deraco M, Baratti D, Kusamura S, Elias D, Glehen O, et al. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for malignant peritoneal mesothelioma: multi-institutional experience. J Clin Oncol. 2009;27(36):6237–42.
Deraco M, Baratti D, Hutanu I, Bertuli R, Kusamura S. The role of perioperative systemic chemotherapy in diffuse malignant peritoneal mesothelioma patients treated with cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Ann Surg Oncol. 2013;20(4):1093–100.
de Boer NL, van Kooten JP, Damhuis RAM, Aerts J, Verhoef C, Madsen EVE. Malignant peritoneal mesothelioma: patterns of care and survival in the netherlands: a population-based study. Ann Surg Oncol. 2019;26(13):4222–8.
Yap TA, Aerts JG, Popat S, Fennell DA. Novel insights into mesothelioma biology and implications for therapy. Nat Rev Cancer. 2017;17(8):475–88.
Hung YP, Dong F, Torre M, Crum CP, Bueno R, Chirieac LR. Molecular characterization of diffuse malignant peritoneal mesothelioma. Mod Pathol. 2020. https://doi.org/10.1038/s41379-020-0588-y.
Shrestha R, Nabavi N, Lin YY, Mo F, Anderson S, Volik S, et al. BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. Genome Med. 2019;11(1):1–12.
Offin M, Yang SR, Egger J, Jayakumaran G, Spencer RS, Lopardo J, et al. Molecular characterization of peritoneal mesotheliomas. J Thorac Oncol. 2022;17(3):455–60.
Dagogo-Jack I, Madison RW, Lennerz JK, Chen KT, Hopkins JF, Schrock AB, et al. Molecular characterization of mesothelioma: impact of histologic type and site of origin on molecular landscape. JCO Precis Oncol. 2022;6: e2100422.
Hiltbrunner S, Fleischmann Z, Sokol ES, Zoche M, Felley-Bosco E, Curioni-Fontecedro A. Genomic landscape of pleural and peritoneal mesothelioma tumours. Br J Cancer. 2022. https://doi.org/10.2139/ssrn.4060087.
Massard C, Michiels S, Ferté C, Le Deley MC, Lacroix L, Hollebecque A, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586–95.
Pam KM, Susan H, Suanna SB, Elizabeth G-M, Ajjai A, Katherine AJ, et al. Rationale and design of the targeted agent and profiling utilization registry study. JCO Precis Oncol. 2018;2:1–14.
Sicklick JK, Kato S, Okamura R, Schwaederle M, Hahn ME, Williams CB, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019;25(5):744–50.
Inc. FM. FoundationOne CDx Technical Information [Available from: https://www.rochefoundationmedicine.com/f1cdxtech.
Hung YP, Dong F, Watkins JC, Nardi V, Bueno R, Dal Cin P, et al. Identification of ALK rearrangements in malignant peritoneal mesothelioma. JAMA Oncol. 2018;4(2):235–8.
Harmon RL, Sugarbaker PH. Prognostic indicators in peritoneal carcinomatosis from gastrointestinal cancer. Int Semin Surg Oncol. 2005;2(1):3.
Dietz MV, van Kooten JP, Paats MS, Aerts JGVJ, Verhoef C, Madsen EVE, et al. Molecular alterations and potential actionable mutations in peritoneal mesothelioma: a scoping review of high-throughput sequencing studies. ESMO Open. 2023. https://doi.org/10.1016/j.esmoop.2023.101600.
Hmeljak J, Sanchez-Vega F, Hoadley KA, Shih J, Stewart C, Heiman D, et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8(12):1548–65.
You JF, Buhard O, Ligtenberg MJ, Kets CM, Niessen RC, Hofstra RM, et al. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br J Cancer. 2010;103(12):1840–5.
Dedeurwaerdere F, Claes KBM, Van Dorpe J, Rottiers I, Van der Meulen J, Breyne J, et al. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci Rep. 2021;11(1):12880.
Tae Hee H, Hongui C, Joon Ho S, Boram L, Jongsuk C, Chung L, et al. Clinical advantage of targeted sequencing for unbiased tumor mutational burden estimation in samples with low tumor purity. J Immunother Cancer. 2020;8(2): e001199.
Arulananda S, Thapa B, Walkiewicz M, Zapparoli GV, Williams DS, Dobrovic A, et al. Mismatch repair protein defects and microsatellite instability in malignant pleural mesothelioma. J Thorac Oncol. 2018;13(10):1588–94.
Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;1:1–15.
Shao C, Li G, Huang L, Pruitt S, Castellanos E, Frampton G, et al. Prevalence of high tumor mutational burden and association with survival in patients with less common solid tumors. JAMA Netw Open. 2020;3(10):e2025109-e.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–20.
Baas P, Scherpereel A, Nowak AK, Fujimoto N, Peters S, Tsao AS, et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. The Lancet. 2021;397(10272):375–86.
Consortium APG. AACR Project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–31.
Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–7.
Panou V, Gadiraju M, Wolin A, Weipert CM, Skarda E, Husain AN, et al. Frequency of germline mutations in cancer susceptibility genes in malignant mesothelioma. J Clin Oncol. 2018;36(28):2863–71.
Pilarski R, Carlo M, Cebulla C, Abdel-Rahman M. BAP1 Tumor Predisposition Syndrome. 1993.
Louie BH, Kurzrock R. BAP1: not just a BRCA1-associated protein. Cancer Treat Rev. 2020;90: 102091.
Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A. 2014;111(1):285–90.
Ismail IH, Davidson R, Gagné JP, Xu ZZ, Poirier GG, Hendzel MJ. Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 2014;74(16):4282–94.
Mateo J, Porta N, Bianchini D, McGovern U, Elliott T, Jones R, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21(1):162–74.
Srinivasan G, Sidhu GS, Williamson EA, Jaiswal AS, Najmunnisa N, Wilcoxen K, et al. Synthetic lethality in malignant pleural mesothelioma with PARP1 inhibition. Cancer Chemother Pharmacol. 2017;80(4):861–7.
Parrotta R, Okonska A, Ronner M, Weder W, Stahel R, Penengo L, et al. A novel BRCA1-associated protein-1 isoform affects response of mesothelioma cells to drugs impairing BRCA1-mediated DNA repair. J Thorac Oncol. 2017;12(8):1309–19.
Borchert S, Wessolly M, Schmeller J, Mairinger E, Kollmeier J, Hager T, et al. Gene expression profiling of homologous recombination repair pathway indicates susceptibility for olaparib treatment in malignant pleural mesothelioma in vitro. BMC Cancer. 2019;19(1):108.
LaFave LM, Béguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21(11):1344–9.
Zauderer MG, Szlosarek PW, Le Moulec S, Popat S, Taylor P, Planchard D, et al. EZH2 inhibitor tazemetostat in patients with relapsed or refractory, BAP1-inactivated malignant pleural mesothelioma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2022;23(6):758–67.
Guazzelli A, Meysami P, Bakker E, Demonacos C, Giordano A, Krstic-Demonacos M, et al. BAP1 status determines the sensitivity of malignant mesothelioma cells to gemcitabine treatment. Int J Mol Sci. 2019;20(2):429.
Okonska A, Bühler S, Rao V, Ronner M, Blijlevens M, van der Meulen-Muileman IH, et al. Functional genomic screen in mesothelioma reveals that loss of function of BRCA1-associated protein 1 induces chemoresistance to ribonucleotide reductase inhibition. Mol Cancer Ther. 2020;19(2):552–63.
Petrilli AM, Fernández-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35(5):537–48.
Curto M, McClatchey AI. Nf2/Merlin: a coordinator of receptor signalling and intercellular contact. Br J Cancer. 2008;98(2):256–62.
Xiao G-H, Gallagher R, Shetler J, Skele K, Altomare DA, Pestell RG, et al. The NF2 tumor suppressor gene product, merlin, inhibits cell proliferation and cell cycle progression by repressing cyclin D1 expression. Mol Cell Biol. 2005;25(6):2384–94.
López-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG. Loss of the tumor suppressor gene NF2, encoding merlin, constitutively activates integrin-dependent mTORC1 signaling. Mol Cell Biol. 2009;29(15):4235–49.
Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338(6104):221.
Ganesan P, Moulder S, Lee JJ, Janku F, Valero V, Zinner RG, et al. Triple-negative breast cancer patients treated at MD Anderson Cancer Center in phase I trials: improved outcomes with combination chemotherapy and targeted agents. Mol Cancer Ther. 2014;13(12):3175–84.
Ou SH, Moon J, Garland LL, Mack PC, Testa JR, Tsao AS, et al. SWOG S0722: phase II study of mTOR inhibitor everolimus (RAD001) in advanced malignant pleural mesothelioma (MPM). J Thorac Oncol. 2015;10(2):387–91.
Cooper J, Xu Q, Zhou L, Pavlovic M, Ojeda V, Moulick K, et al. Combined inhibition of NEDD8-activating enzyme and mTOR suppresses NF2 loss-driven tumorigenesis. Mol Cancer Ther. 2017;16(8):1693–704.
van Janse Rensburg HJ, Yang X. Essential signaling in NF2 loss-related tumours: the therapeutic potential of CRL4(DCAF1) and mTOR combined inhibition. J Thorac Dis. 2017;9(10):3533–6.
Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–7.
Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-Repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697–708.
de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091–102.
Bang YJ, Im SA, Lee KW, Cho JY, Song EK, Lee KH, et al. Randomized, double-blind phase II trial with prospective classification by ATM protein level to evaluate the efficacy and tolerability of olaparib plus paclitaxel in patients with recurrent or metastatic gastric cancer. J Clin Oncol. 2015;33(33):3858–65.
Olson D, Bhalla S, Yang X, Martone B, Kuzel TM. Novel use of targeted therapy via PARP-inhibition in a rare form of papillary renal cell carcinoma: a case report and literature review. Clin Genitourin Cancer. 2016;14(4):e445–8.
Fennell DA, King A, Mohammed S, Branson A, Brookes C, Darlison L, et al. Rucaparib in patients with BAP1-deficient or BRCA1-deficient mesothelioma (MiST1): an open-label, single-arm, phase 2a clinical trial. Lancet Respir Med. 2021;9(6):593–600.
Acknowledgements
Not applicable.
Funding
NGS by use of the Foundation Medicine (FMI) F1CDx platform was financially supported by Roche.
Author information
Authors and Affiliations
Contributions
JK and MD contributed equally to this manuscript. The study concept and design were provided by CV, JA, EM, and JT. MD and JP drafted the manuscript. JP provided the clinical data. JT and HD provided the pathological data. All authors have reviewed and revised the manuscript and approved the submission.
Corresponding author
Ethics declarations
Conflict of interest
NGS by use of the Foundation Medicine (FMI) F1CDx platform was financially supported by Roche. The authors report no further competing interests.
Ethics approval and consent to participate
The study was performed according to the principles of the Declaration of Helsinki and was approved by the EMC local ethics committee (MEC 2018–1286).
Consent for publication
Patients provided written permission to use this tissue for research purposes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van Kooten, J.P., Dietz, M.V., Dubbink, H.J. et al. Genomic characterization and detection of potential therapeutic targets for peritoneal mesothelioma in current practice. Clin Exp Med 24, 80 (2024). https://doi.org/10.1007/s10238-024-01342-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10238-024-01342-y