Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease with a poor prognosis. The current risk stratification system is essential but remains insufficient to select the best schedules. Cysteine-rich protein 1 (CSRP1) is a member of the CSRP family and associated with poor clinicopathological features in many tumors. This study aimed to explore the clinical significance and molecular mechanisms of cysteine- and glycine-rich protein 1 (CSRP1) in AML. RT-qPCR was used to detect the relative expression of CSRP1 in our clinical cohort. Functional enrichment analysis of CSRP1-related differentially expressed genes was carried out by GO/KEGG enrichment analysis, immune cell infiltration analysis, and protein–protein interaction (PPI) network. The OncoPredict algorithm was implemented to explore correlations between CSRP1 and drug resistance. CSRP1 was highly expressed in AML compared with normal samples. High CSRP1 expression was an independent poor prognostic factor. Functional enrichment analysis showed neutrophil activation and apoptosis were associated with CSRP1. In the PPI network, 19 genes were present in the most significant module, and 9 of them were correlated with AML prognosis. The high CSRP1 patients showed higher sensitivity to 5-fluorouracil, gemcitabine, rapamycin, cisplatin and lower sensitivity to fludarabine. CSRP1 may serve as a potential prognostic marker and a therapeutic target for AML in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous and highly aggressive hematologic malignancy [1]. It is the most prevalent form of acute leukemia in adults, with an annual incidence of 4.3/100,000 and an increasing incidence risk with older age [2]. Existing standard treatments include intensive chemotherapy and allogeneic hematopoietic stem cell transplantation (HSCT). Since 2017, the U.S. Food and Drug Administration (FDA) has approved eight new drugs for the treatment of AML, including the FLT3 inhibitors midostaurin and gicitinib, the IDH inhibitors efusitinib and enalcitinib, the anti-CD33 monoclonal antibody gemtuzumab ozogamicin, CPX351, the hedgehog pathway inhibitor glasdegib, and BCL-2 inhibitor venetoclax. Targeted therapies are more effective and less toxic than the conventional chemotherapy [3]. Despite significant advances in these new therapies, primary and secondary resistance remains a major obstacle in treating AML. Therefore, pre-evaluation of the likelihood of resistance is vital for treatment options [4, 5]. Allogeneic HSCT has become an effective tool for AML cure by preventing AML relapse through high-dose chemotherapy and graft-versus-leukemia effects. However, how to screen the population for transplantation is still a challenge in AML treatment. According to the current cytogenetic risk stratification, patients are classified into three risk groups (favorable group, intermediate group, and adverse group). However, patients classified as favorable or intermediate group still have high relapse and drug resistance rates, suggesting that the current risk stratification system is still inadequate. Therefore, the creation of novel and trustworthy prognostic biomarkers for AML is urgently required.
Cysteine-rich protein 1 (CSRP1) is a member of the CSRP family. This gene family contains a group of LIM domain proteins, which are proposed to be involved in regulatory processes essential for development and cellular differentiation. CSRP1 is located on human chromosome 1q32.1 [6]. The LIM protein, CRP1, is a general marker for smooth muscle lineages [7]. CRP1 localizes to the nucleus and the cytoplasm with different functions depending on its location. When CRP1 is in the nucleus, it regulates interactions between transcription factors and promotes the upregulation of smooth muscle-specific genes [8]. When CRP1 is in the cytoplasm, it localizes to the adhesion patch and the actin cytoskeleton to regulate actin filament bundles [9].
Only a few reports have been made about CSRP1's connection to cancer thus far. CSRP1 is associated with poor clinicopathological features in adrenocortical carcinoma (ACC) [10]. Hepatocellular carcinoma (HCC) causes abnormal methylation to inactivate CSRP1, which may be a key biomarker for cancer [11]. CSRP1 was used to forecast when benign prostatic hyperplasia will turn into prostate cancer and may have an effect on disease-free survival [12]. In addition, CSRP1 is also associated with colorectal cancer [13] and breast cancer [14].
There has been no research to investigate the expression profile and function of CSRP1 in AML. In this study, we examined the database and our cohort to investigate the expression of CSRP1 in AML and its prognostic significance. We also studied the differentially expressed genes related to CSRP1 expression and explored their potential roles in AML through GO and KEGG enrichment, immune infiltration, protein interaction analysis, and drug sensitivity analysis.
Materials and methods
Patients and treatment regimen
Bone marrow samples from 224 newly diagnosed AML patients and 23 healthy individuals were obtained at the First Affiliated Hospital of Zhengzhou University between February 2017 and July 2020. We named our cohort as ZZU cohort. The inclusion criteria included: 1) patients were diagnosed as AML in our hospital; 2) patients were ≥ 14 years old; and 3) patients accepted at least one course of chemotherapy in our hospital. The exclusion criteria were patients who didn’t treat or treat elsewhere and patients with acute promyelocytic leukemia. Patient clinical information, such as age, gender, risk stratification, FAB classification, gene mutations, and other basic information, was gathered from electronic medical records. All data were collected independently by two researchers who received training to reliably complete all assessments. Details of treatment regimens are reported [15, 16]. Induction chemotherapy regimens include IA and DA regimens: standard-dose cytarabine (Ara-C) 100–200 mg·m−2·d−1 × 7 d combined with idarubicin 10–12 mg·m−2·d−1 × 3d or daunorubicin 60 mg·m−2·d−1 × 3d. The treatment plan after remission is high-dose Ara-C (2–3 g/m2, once every 12 h, 3d). Those who have not undergone HSCT share four courses; those who receive HSCT share two courses, followed by HSCT. For elderly patients over 60 years old or patients who cannot tolerate intensive chemotherapy, they received chemotherapy with demethylating drugs ± CAG ± venetoclax regimen until progression. In total, 38 subjects received an allogeneic HSCT. Subjects were followed up until death, loss to follow-up or December 2022. The follow‐up was conducted through in‐clinic follow‐up and telephone calls. Patients were classified into low CSRP1 group and high CSRP1 group according to the expression of CSRP1 at diagnosis. The clinical significance of CSRP1 in the AML cohort was analyzed retrospectively. Complete remission, relapse, risk stratification, overall survival (OS), and relapse-free survival (RFS) were defined as described according to the 2017 ELN recommendations from an international expert panel [17]. The study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, and written informed consent was exempted by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
RNA extraction, cDNA synthesis and RT-qPCR
Bone marrow mononuclear cells were obtained via density gradient centrifugation. Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). The cDNA was synthesized using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) (18). CSRP1 transcript levels were detected by the Taqman method using real-time quantitative polymerase chain reaction (RT-qPCR) as previously described [18]. Serial dilutions of plasmids expressing CSRP1 and ABL1 were amplified to construct standard quantification curves [19]. The CSRP1 and ABL1 copy numbers were calculated from standard curves. As previously described, the CSRP1 transcript levels were calculated as the ratio of the CSRP1 copy number/ABL1 copy number [20]. The primers and probe sequences are shown in Table S1.
Data acquisition
We obtained clinical information and gene expression profiles for AML patients from three independent sources. The first source is Tyner's study, which included 405 AML patients (hereafter referred to as the Beat-AML dataset) [21]. The second source is the TCGA database, which provides RNA-seq expression data and detailed clinical data on 173 AML patients (hereafter referred to as the TCGA-LAML dataset) [22]. The third source is the GSE12417 from the GEO database (https://www.ncbi.nlm.nih.gov/geo/; hereafter referred to as the GSE12417 dataset) [23]. The AML samples' Level 3 HTSeq-FPKM and HTSeq-Count data were downloaded from the TCGA website (https://portal.gdc.cancer.gov/repository) for further investigation.
Survival analyses
We utilized the pROC package to determine the optimal cutoff value for each gene and divided AML patients into two subgroups: the “high-expression” and “low-expression” groups according to the cutoff value of the gene. Then we performed Kaplan–Meier survival analysis to investigate the prognostic value of gene expression using the survival package.
Differentially expressed gene (DEG) analysis
The DESeq2 R package was used to compare the expression data of low and high expression of CSRP1 (cutoff value of 50%) in AML samples to identify DEGs [24]. Heatmap analysis of the top 20 DEGs was performed.
Functional enrichment analysis of DEGs
Functional enrichment analysis was performed on DEGs with |logFC| > 0.5 and padj < 0.05. The ClusteProfiler package [25] in R was used to implement Gene Ontology (GO) functional analysis, which included cellular component (CC), molecular function (MF), and biological process (BP) analysis, as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Analysis of immune infiltration using single-sample gene set enrichment analysis (ssGSEA)
CSRP1 immune infiltration analysis was carried out using ssGSEA utilizing the GSVA package [26] in R. As previously stated [27], 24 kinds of infiltrating immune cells were obtained. The Spearman correction was used to examine the relationship between CSRP1 and the enrichment scores of 24 different kinds of immune cells. The Wilcoxon rank-sum test was used to examine the enrichment scores of the groups with high and low CSRP1 expression.
The protein–protein interaction (PPI) network
The Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org) was used to predict the DEG PPI network [28]. The cutoff criterion was set at an interaction score level of 0.4. The PPI network was mapped with Cytoscape (version 3.6.1) [29], and the most critical modules in the PPI network were identified with MCODE (version 1.5.1) [30]. The following were the selection criteria: MCODE scores greater than 5, degree cutoff = 2, node score cutoff = 0.2, Max depth = 100, and k-score = 2.
Generation and prediction of prognostic models
A nomogram was developed using the rms R package (version 6.2–0) to individualize the prediction of overall survival (OS) in AML patients, which comprised key clinical features and calibration plots. The calibration curves were visually assessed by plotting the nomogram-predicted probabilities against the observed rates, with the 45 line representing the best predictive values. All statistical tests were two-tailed, and the statistical significance threshold was set at 0.05.
Chemotherapy response prediction
The oncoPredict package of R was used to forecast medication sensitivity to 5-fluorouracil, gemcitabine, rapamycin, cisplatin, and fludarabine with the goal of enhancing tailored therapy. Accordingly, drug sensitivity values (measured by AUC, the area under the concentration–response curve) were estimated followed by a comparison of the values between the high and low CSRP1 groups. High AUC means low sensitivity.
Statistical investigation
R (3.6.3) was used to perform and show all statistical analyses and figures. Wilcoxon rank-sum test was used to evaluate CSRP1 expression in unpaired samples, while Wilcoxon signed-rank test was utilized in paired samples. Quantitative data are presented as means ± SD (standard deviations) or medians with interquartile ranges. To assess the connection between clinical/cytogenetic features and CSRP1 expression, the Kruskal–Wallis test, Wilcoxon signed-rank test, and logistic regression analysis were performed. The prognostic variables were evaluated using Cox regression analysis and the Kaplan–Meier technique. The cutoff of CSRP1 was calculated by X-tile. There were few missing data on the key variables in our main analysis. Participants who were lost to follow-up were deemed alive after loss to follow-up. Multivariate analyses were conducted, controlling for confounders.
Results
Overexpression of CSRP1 in AML and expression of CSRP1 in pan-cancers
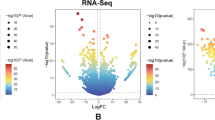
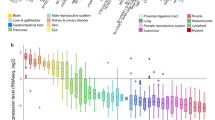
RNA-seq data were obtained in the TCGA and GTEx databases. By comparing the expression of CSRP1 in normal samples in the GTEX database and corresponding tumor samples in the TCGA database, CSRP1 was found to be significantly overexpressed in 7 cancers, including acute myeloid leukemia (LAML), and underexpressed in 18 cancers (Fig. 1A). For the CSRP gene family, CSRP1 was significantly overexpressed in AML, while CSRP3 was significantly reduced in AML compared to healthy controls (Fig. 1B). Furthermore, by comparing the expression of CSRP1 in 23 normal bone marrow (NBM) and 224 newly diagnosed AML patients in the ZZU cohort, we confirmed that CSRP1 was overexpressed in AML (Fig. 1C).
AML samples showed a higher expression of CSRP1 compared to normal samples. A Expression levels of CSRP1 in paired samples of normal and tumor patients in different cancers. B Comparisons of gene expressions among different CSRP family for normal and AML samples. C AML samples from the ZZU cohort showed a significant increase in CSRP1 expression compared to normal bone marrow samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001
Association between CSRP1 expression and clinical features
The main clinical characteristics of AML in the ZZU cohort are shown in Table 1. A total of 224 AML patients were included in the prognostic analysis. The median follow-up time was 529 days. Up to the follow-up time, 138 patients died. The AML patients were divided into the low CSRP1 group (109 cases) and the high CSRP1 group (115 cases) according to the cut-off CSRP1 expression (225%). The high CSRP1 group had higher BM-blasts and more DNMT3A mutations than the low CSRP1 group (P < 0.05, Table 1 and Fig. 5G). We also compared the expression of CSRP1 in AML patients by the other characteristics. The expression levels of CSRP1 showed no significant difference in patients with different gender, age, WBC, cytogenetic risk group, BM-blasts, and PB-blasts (P > 0.05; Fig. 2A–F).
Association between CSRP1 expression and clinical features in the ZZU cohort. A–F Comparisons of CSRP1 expression in AML patients based on gender A, age B, cytogenetics risk C, WBC D, BM-blasts E, PB-blasts F. G Association among CSRP1 expression, survival status and common gene mutations in AML. ns: P > 0.05; *, P < 0.05
High CSRP1 impacted the prognosis of AML
Patients with high CSRP1 expression had a significantly worse prognosis than those with low CSRP1 expression (hazard ratio [HR], 2.36 (1.53–3.64); P < 0.001; Fig. 3A) in the TCGA-LAML dataset. The predictive significance of elevated CSRP1 was verified in the Beat-AML dataset (Fig. 3B), the ZZU cohort (Fig. 3C), and the GSE12417 dataset (Fig. 3D–E). The time-dependent ROC curve from the TCGA-LAML dataset demonstrated that CSRP1 was an excellent predictor of AML patient survival.
High expression of CSRP1 was associated with poor OS in AML patients. A–E Kaplan–Meier curves of OS in the TCGA-LAML dataset A, the Beat-AML dataset B, the ZZU cohort C, the GSE12417-GPL96 dataset D, and the GSE12417-GPL570 dataset E and F Time-dependent ROC curve of CSRP1 in TCGA-LAML dataset. OS, overall survival; HR, hazards ratio
Based on the TCGA-LAML dataset, independent prognostic factors for worse OS included CSRP1 (high vs. low, P < 0.001), cytogenetic risk (adverse vs. favorable, P < 0.001; intermediate vs. favorable, P = 0.002), and age (> 60 vs. ≤ 60, P < 0.001) (Table 2 and Fig. 4A). The independent prognostic value of these three factors was further confirmed in the ZZU cohort (Fig. 4B).
Construction of the nomogram model for AML patients. A–B The forest plots showed that age, cytogenetic risk, and CSRP1 expression are independent factors for poor prognosis in the TCGA-LAML dataset A and the ZZU cohort B. C–D Nomogram for predicting the probability of 1-, 2-, 3-year OS for AML in the C TCGA-LAML dataset and D the ZZU cohort. E–F Calibration plot of the nomogram for predicting the probability of OS at 1, 2, and 3 years for AML in the E TCGA-LAML dataset and F the ZZU cohort
Prognostic model of CSRP1 in AML
A nomogram was developed based on the Cox regression analyses in the TCGA dataset (Fig. 4C) and the ZZU cohort (Fig. 4D) to better predict AML patients' prognoses. Three independent prognostic variables, age, cytogenetic risk, and CSRP1 expression, were included in the model at a statistical significance level of 0.05. A point scale was utilized to allocate points to these factors based on multivariate Cox analysis. Results from the nomogram calibration curve of OS prediction were consistent with observations of all patients in both the TCGA dataset (Fig. 4E) and the ZZU cohort (Fig. 4F).
Identification of DEGs in AML samples with low and high CSRP1 expression
The gene expression profiles of the high and low CSRP1 groups were analyzed for differences in median mRNA expression. A total of 2758 DEGs from RNA-seq-HTSeq counts were found to be statistically significant between the high and low CSRP1 groups (|log fold change (logFC)|> 0.5, P adj < 0.05). (Fig. 5A). The heat map depicted the top ten up-regulated DEGs and top ten down-regulated DEGs between the high and low CSRP1 groups (Fig. 5B).
GO/KEGG enrichment analysis of DEGs comparing patients with high or low CSRP1 expression in the TCGA-LAML dataset. A Volcano map of the DEGs, including 862 down-regulated genes and 1896 up-regulated genes. B Heat map showing the top ten up-regulated and the top ten down-regulated genes. The samples are shown on the X-axis, while the DEGs are shown on the Y-axis. C–D GO enrichment analysis of the up-regulated DEGs. MF, molecular function. CC, cellular component. BP, biological process. D KEGG enrichment analysis of the up-regulated DEGs. Different categories were shown on the Y-axis, while the X-axis reflected the percentage of DEGs
Functional enrichment analysis of DEGs
Biological process (BP) associated with high CSRP1 included neutrophil activation, neutrophil activation involved in immune response, neutrophil degranulation, and neutrophil mediated immunity (Fig. 5C). Cellular components (CC) associated with high CSRP1 included secretory granule membrane, ficolin-1-rich granule, tertiary granule, and cell leading edge (Fig. 5C). Molecular function (MF) associated with high CSRP1 included actin binding, Rho GTPase binding, cytokine binding, and cytokine receptor activity (Fig. 5C). KEGG included phagosome, cytokine-cytokine receptor interaction, lysosome, oxytocin signaling pathway, cell adhesion molecules, rap1 signaling pathway, chemokine signaling pathway, gap junction, HIF-1 signaling pathway, proteoglycans in cancer, Fc gamma R-mediated phagocytosis, natural killer cell mediated cytotoxicity, JAK-STAT signaling pathway, and apoptosis (Fig. 5D).
The prognostic value of the top ten up-regulated DEGs and top ten down-regulated DEGs was further investigated. Six of the top ten DEGs that were up-regulated (ONECUT2, CES1, CD163, KCNN2, THBS1, and B3GALT1) and six of the top ten DEGs that were down-regulated (MYT1L, ASTN1, DLL3, MYO18B, POU4F1, and PCDH10) also had vital prognostic significance in the TCGA AML dataset (Fig. 6).
Prognostic value of DEGs in TCGA-LAML dataset. A–F Six of the top ten up-regulated DEGs (ONECUT2 A, CES1 B, CD163 C, KCNN2 D, THBS1 E, and B3GALT1 F) showed an adverse effect on OS. (G–L). Six of the top ten down-regulated DEGs (MYT1L G ASTN1 H, DLL3 I, MYO18B J, POU4F1 K, and PCDH10 L) had a protective effect on OS
Analysis of immune infiltration in AML
CSRP1 expression in the AML microenvironment was associated with the level of immune cell infiltration as measured by ssGSEA. Mainly, CSRP1 was negatively related to T cells and T helper cells and positively associated with macrophages (Fig.S1C), neutrophils (Fig.S1D), NK CD56 dim cells, NK CD56 bright cells, Tem, and iDC, TFH, Th17 cells, Th1 cells, eosinophils, Treg, and aDC (Fig.S1A). Patients with high CSRP1 had higher enrichment scores of aDC, Eosinophils, iDC, macrophages, neutrophils, NK CD56 bright cells, NK CD56 dim cells, Tem, Th1 cells, Treg, and lower enrichment scores of T helper cells.
PPI enrichment analysis in AML and validation of hub genes
The PPI network was constructed using the String website and the data were generated by Cytoscape (MCODE plug-in). A total of 321 DEGs were imported into the PPI network. Complete module including all genes is presented in Fig.S2A. We obtained 235 nodes and 900 edges. Nineteen genes were present in the most significant module (MCODE score: 7.667; Fig.S2B). The TCGA-LAML dataset was then used for OS analysis on the hub genes. Nine of the 19 hub genes, including CD163, CX3CR1, C5AR1, THSD7A, ADMATS18, IL10, THBS1, ADAMTS15, and LILRB2, were correlated with OS in AML (P < 0.05, Fig.S3).
CSRP1 expression can predict sensitivity to the common chemotherapy agents in AML treatment
We utilized the “oncoPredict” tool to estimate sensitivity to frequently used chemotherapy agents to better correlate the CSRP1 expression with clinical practice. Accordingly, drug sensitivity of patients in high and low CSRP1 groups to multiple chemotherapy agents including 5-fluorouracil, gemcitabine, rapamycin, cisplatin, and fludarabine was predicted. Based on the findings, the high CSRP1 groups of patients in the TCGA datasets showed higher sensitivity to 5-fluorouracil, gemcitabine, rapamycin, and cisplatin and lower sensitivity to fludarabine (Fig.S4). The relationship of sensitivity to other chemotherapy agents with CSRP1 expression is listed in the supplementary data.
Discussion
In this study, we explored the relationship between CSRP1, the clinicopathological features and prognosis of AML. We found that CSRP1 was highly expressed in adult AML, which was associated with a higher proportion of bone marrow blasts, a higher frequency of DNMT3A mutation and a poor prognosis in AML patients. In addition, we constructed a nomogram to predict OS for AML based on age, cytogenetic risk stratification, and CSRP1 expression levels and explored the possible mechanisms of CSRP1 function.
Through pan-cancer analysis, we found that, unlike other tumor markers that are always highly or lowly expressed in different tumors, CSRP1 shows different expression patterns between different tumors, with CSRP1 highly expressed in 7 cancers and lowly expressed in 18 tumors. It suggests that CSRP1 has a complex mechanism of regulation and can function as either an oncogene or oncogene suppressor in different cancer species or under different circumstances. In addition, the expression of CSRP gene family in AML is inconsistent, with the CSRP1 highly expressed and CSRP3 lowly expressed in AML. We have systematically investigated the role of the CSRP2 gene in AML and found that its low expression is associated with poor prognosis in AML [15]. At the same time, the knockdown of CSRP2 promotes proliferation and cycle progression in AML cell lines [15]. In contrast, no studies on CSRP1 in AML have been reported. We confirmed the high expression of CSRP1 in AML by comparing 224 adult AML patients with 23 healthy controls, which is consistent with the database results. All of these suggest that CSRP1 may play an essential role in AML.
Next, we investigated the relationship between CSRP1 gene expression, the clinicopathological features, and gene mutations using data from 224 adult AML cases. CSRP1 expression did not correlate with gender, age, risk stratification, or WBC at diagnosis. The analysis of baseline data showed a higher proportion of bone marrow blasts and a higher frequency of DNMT3A mutations in the high CSRP1 group. DNMT3A is one of the most frequently mutated genes in AML [31] and is an independent prognostic factor used for risk stratification [32]. It may suggest a higher tumor burden and a higher incidence of adverse prognostic mutations in those with high CSRP1 expression.
We next explored the impact of CSRP1 gene expression levels on overall survival in adult AML patients. We found that high CSRP1 expression was associated with poor OS through multiple adult AML database cohort studies, including TCGA-LAML, Beat-AML, and GEO databases. Moreover, we further validated this result with the ZZU cohort. To further optimize the current stratification system for AML and facilitate clinical application, we first performed a univariate analysis. Initial screening revealed that age > 60 years old, worse cytogenetic risk stratification and high CSRP1 expression were independent poor prognostic factors. The prognostic significance of age and karyotype stratification is well established. To facilitate clinical application, we further developed a nomogram and applied a calibration plot to validate the model. This model performed well for both the TCGA-LAML dataset and the ZZU cohort. This finding is beneficial for further optimizing the stratification system, especially for some patients classified as intermediate risk according to the current stratification, who can now better evaluate transplantation or chemotherapy according to the current guideline recommendation. This stratification system can further score, stratify and guide the treatment selection.
The overexpression of CSRP1 and its associated poor prognostic value in AML suggest that it may play a role in AML. To further explore the mechanism of action of CSRP1, we subjected patients with high and low CSRP1 expression to differential gene expression analysis, followed by GO and KEGG functional enrichment. GO-BP enrichment analysis revealed that CSRP1 was closely associated with neutrophil function, which was confirmed in subsequent microenvironmental correlation analysis. Upregulating neutrophil elastase (NE) promoted the growth of leukemia cells and decreased the proportion of apoptotic cells [33]. GO-BP enrichment analysis reveals that CSRP1 is associated with actin binding and Rho GTPs binding. AML with FLT3-ITD mutations is characterized by RAC1-dependent actin cytoskeleton remodeling that substantially contributes to the acquisition of resistance to midostaurin in vitro [34]. Yang et al. [35] analyzed the expression patterns and prognostic significance of Rho family GTPases in AML and found that RhoBTB3 was significantly downregulated in AML bone marrow compared to healthy controls and correlated with prognosis of AML. KEGG enrichment analysis revealed that CSRP1 was associated with cell adhesion molecules, rap1 signaling pathway, HIF-1 signaling pathway, JAK-STAT signaling pathway, and apoptosis. Cellular adhesion molecules also impact the poor prognosis of AML and may be used as targets for AML-specific therapies [36]. Also, the Rap1 signaling pathway plays a crucial role in cancer [37]. HIF signaling has been implicated in myeloid cell survival, and PI3K/Akt is known to induce HIF-1 transcription [38]. The signaling pathway JAK/STAT plays a critical role in the development and progression of AML [39].
In the PPI analysis, nine out of the 19 hub genes (CD163, CX3CR1, C5AR1, THSD7A, ADMATS18, IL10, THBS1, ADAMTS15, and LILRB2) were correlated with OS in AML. Notably, CD163 and THBS1 were among the 20 genes screened for the most significant differences based on CSRP1 expression levels. CD163 was a specific marker for macrophages of M2 type and was identified as a potential target for the therapeutic intervention of AML [40, 41]. The association of CSRP1 with poor prognosis and macrophages in AML may be acting through CD163, but this requires further experimental confirmation. THBS1 is a novel serum prognostic factors of AML [42]. CX3CR1 was identified as one of the distinctive features of AML cells for universal MRD monitoring [43]. For AML patients who are ineligible for standard treatment with chemotherapy and HSCT and who also experience less severe side effects, Laura Jimbu et al. propose that manipulation of both the co-inhibitory network (with anti-PD-L1 blocking antibodies) and suppressor network (with anti-IL-10 blocking antibodies) is an appealing immunotherapeutic intervention. To increase the therapeutic effectiveness of treating AML illness, the suggested combination of these two immunotherapies provides a novel strategy that can be easily used in the clinic [44]. Endothelial cells (ECs)-derived small extracellular vesicles contained a high level of ANGPTL2, which accelerated leukemia progression via binding to the LILRB2 receptor [45]. The other genes, including C5AR1, THSD7A, ADAMTS18, and ADAMTS15, have not yet been reported in AML-related studies and also require further investigation.
Finally, we also aimed to investigate whether CSRP1 expression is capable of prognosticating the patients’ response to the commonly used chemotherapeutic agents. Our findings revealed that patients with higher CSRP1 expression were more sensitive to certain chemotherapy agents including 5-fluorouracil, gemcitabine, rapamycin, and cisplatin and more resistant to fludarabine. This will help us in the selection of drugs in case of relapse and refractory patients.
In summary, CSRP1 was highly expressed in adult AML, and such high-level expression of CSRP1 was related to a poor prognosis in adult AML. CSRP1 may serve as a potential prognostic marker and a therapeutic target for AML in the future. Further verification is expected to be carried out to reveal the clinical significance and biological impacts of CSRP1 in AML.
Data availability
The datasets generated and analyzed during the current study are not publicly available due to patient privacy considerations, but are available from the corresponding author on reasonable request (Shujuan Wang: fccwangsj1@zzu.edu.cn).
References
Luyao L, Yehuda GA, Zi-Ning L, et al. Genetic biomarkers of drug resistance: a compass of prognosis and targeted therapy in acute myeloid leukemia. Drug Resist Updat. 2020;52:100703.
Mims AS, Kohlschmidt J, Borate U, et al. A precision medicine classification for treatment of acute myeloid leukemia in older patients. J Hematol Oncol. 2021;14(1):96.
Ma J, Ge Z. Recent advances of targeted therapy in relapsed/refractory acute myeloid leukemia. Bosn J Basic Med Sci. 2021;21:96.
Murphy T, Yee KWL. Cytarabine and daunorubicin for the treatment of acute myeloid leukemia. Expert Opin Pharmacother. 2017;18(16):1765–80.
Thomas B, Emilie R, Sylvain T, Frederic L. Advances in treatment formulations for acute myeloid leukemia. Drug Discov Today. 2018;23(12):1936–49.
Erdel M, Weiskirchen R. Assignment1 of CSRP1 encoding the LIM domain protein CRP1, to human chromosome 1q32 by fluorescence in situ hybridization. Cytogenet Genome Res. 1998;83(1–2):10–1.
Henderson JR, Macalma T, Brown D, Richardson JA, Olson EN, Beckerle MC. The LIM protein, CRP1, is a smooth muscle marker. Dev dyn publ Am Assoc Anat. 1999;214(3):229–38.
David FC, Narasimhaswamy SB, Dinakar I, et al. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4(1):107–18.
Thuan CT, Coreyayne S, Tamara SF, Jeffrey AG. Cysteine-rich protein 1 (CRP1) regulates actin filament bundling. BMC Mol Cell Biol. 2005;6(1):45.
Xu F, Zhang P, Yuan M, Yang X, Chong T. Bioinformatic screening and identification of downregulated hub genes in adrenocortical carcinoma. Exp Ther Med. 2020;20(3):2730–42.
Hirasawa Y, Arai M, Imazeki F, et al. Methylation status of genes upregulated by demethylating agent 5-aza-2′-deoxycytidine in hepatocellular carcinoma. Oncology. 2006;71(1–2):77–85.
Chen X, Ma J, Xu C, et al. Identification of hub genes predicting the development of prostate cancer from benign prostate hyperplasia and analyzing their clinical value in prostate cancer by bioinformatic analysis. Discov Oncol. 2022;13(1):54.
Zhou CZ, Qiu GQ, Wang XL, et al. Screening of tumor suppressor genes on 1q31.1–32.1 in Chinese patients with sporadic colorectal cancer. Chin Med J (Engl). 2008;121(24):2479–86.
López-Cortés A, Cabrera-Andrade A, Vázquez-Naya JM, et al. Prediction of breast cancer proteins involved in immunotherapy, metastasis, and RNA-binding using molecular descriptors and artificial neural networks. Sci Rep. 2020;10(1):8515.
Wang S, Zhang Y, Liu Y, et al. Inhibition of CSRP2 promotes leukemia cell proliferation and correlates with relapse in adults with acute myeloid leukemia. Onco Targets Ther. 2020;13:12549–60.
Li M, Liu Y, Liu Y, et al. Downregulation of GNA15 inhibits cell proliferation P38 MAPK pathway and correlates with prognosis of adult acute myeloid leukemia with normal karyotype. Front Oncol. 2021;11:724435.
Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–47.
Shujuan W, Chong W, Weiqiong W, Qianqian H, Yanfang L. High RASD1 transcript levels at diagnosis predicted poor survival in adult B-cell acute lymphoblastic leukemia patients. Leuk Res. 2019;80:26–32.
Gabert J, Beillard E, van der Velden VH, et al. Standardization and quality control studies of “real-time” quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia-a Europe against cancer program. Leukemia. 2003;17(12):2318–57.
Wang SJ, Wang PZ, Gale RP, et al. Cysteine and glycine-rich protein 2 (CSRP2) transcript levels correlate with leukemia relapse and leukemia-free survival in adults with B-cell acute lymphoblastic leukemia and normal cytogenetics. Oncotarget. 2017;8(22):35984–6000.
Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–31.
Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74.
Metzeler KH, Hummel M, Bloomfield CD, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112(10):4193–201.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7.
Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–95.
Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13.
Demchak B, Hull T, Reich M, et al. Cytoscape: the network visualization tool for GenomeSpace workflows. F1000Research. 2014;3:151.
Bandettini WP, Kellman P, Mancini C, MultiContrast, et al. Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson. 2012;14(1):83.
Emma COB, John B, Timothy C. DNMT3A: the DioNysian MonsTer of acute myeloid leukaemia. Ther Adv Hematol. 2014;5(6):187–96.
Zhang Q, Wu X, Cao J, Gao F, Huang K. Association between increased mutation rates in DNMT3A and FLT3-ITD and poor prognosis of patients with acute myeloid leukemia. Exp Ther Med. 2019;18(4):7891.
Jiang K, Ma P, Yang X, et al. Neutrophil elastase and its therapeutic effect on leukemia cells. Mol Med Rep. 2015;12(3):4165–72.
Garitano TA, Sancho A, Götz R, et al. Actin cytoskeleton deregulation confers midostaurin resistance in FLT3-mutant acute myeloid leukemia. Commun Biol. 2021;4(1):799.
Yang S, Liu W, Peng J, et al. High expression of RhoBTB3 predicts favorable chemothrapy outcomes in non-M3 acute myeloid leukemia. J Cancer. 2021;12(14):4229–39.
Shi L, Huang R, Lai Y. Identification and validation of signal recognition particle 14 as a prognostic biomarker predicting overall survival in patients with acute myeloid leukemia. BMC Med Genomics. 2021;14(1):127.
Wu J, Niu Q, Yuan J, Xu X, Cao L. Novel compound cedrelone inhibits hepatocellular carcinoma progression via PBLD and Ras/Rap1. Exp Ther Med. 2019;18(6):4209–20.
Ho PA, Alonzo TA, Kopecky KJ, et al. Molecular alterations of the IDH1 gene in AML: a children’s oncology group and Southwest oncology group study. Leukemia. 2010;24(5):909–13.
Zhu X, Guo Q, Zhu M, et al. HLX affects cell cycle and proliferation in AML cells via the JAK/STAT signaling pathway. Oncol Lett. 2020;20(2):1888–96.
Bachli EB, Schaer DJ, Walter RB, Fehr J, Schoedon G. Functional expression of the CD163 scavenger receptor on acute myeloid leukemia cells of monocytic lineage. J Leukoc Biol. 2006;79(2):312–8.
Hwang I, Kim JW, Ylaya K, et al. Tumor-associated macrophage, angiogenesis and lymphangiogenesis markers predict prognosis of non-small cell lung cancer patients. J Transl Med. 2020;18(1):443.
Zhu L, Li Q, Wang X, et al. THBS1 Is a novel serum prognostic factors of acute myeloid leukemia. Front Oncol. 2019;9:1567.
Coustan-Smith E, Song G, Shurtleff S, et al. Universal monitoring of minimal residual disease in acute myeloid leukemia. JCI insight. 2018;3(9):e98561.
Jimbu L, Mesaros O, Neaga A, et al. The potential advantage of targeting both PD-L1/PD-L2/PD-1 and IL-10–IL-10R pathways in acute myeloid leukemia. Pharm (Basel Switz). 2021;14(11):1105.
Dan H, Guohuan S, Xiaoxin H, et al. ANGPTL2-containing small extracellular vesicles from vascular endothelial cells accelerate leukemia progression. J Clin Investig. 2021;131(1):e138986.
Funding
This work was supported by the National Natural Science Foundation of China [Grant number 81800137], the Priority Academic Program of Henan Higher Education Institutions [Grant number 23A320035], and the Young and Middle-Aged Health Science and Technology Innovation Talent Project of Henan Province [Grant number YXKC2021011].
Author information
Authors and Affiliations
Contributions
QQH and SJW contributed to conception and design; QQH, YL, LYS, YFC, and LY contributed to data acquisition and data analysis/interpretation; ZXJ, YFL, CW, and LS performed project administration; all authors performed final approval of manuscript; QQH, YJL, and SJW performed manuscript drafting or manuscript revision. Also, we thank Medical Transformation Center of the First Affiliated Hospital of Zhengzhou University for the experimental platform provided for this study.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
We thank all the treating physicians for allowing us to enroll their patients and the patients for allowing us to analyze their data. All data comply with the relevant regulations of the Ethics Committee of the First Affiliated Hospital of Zhengzhou University and have been approved for publication. The Ethical Approval number is 2022-KY-0852-001.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hao, Q., Liu, Y., Liu, Y. et al. Cysteine- and glycine-rich protein 1 predicts prognosis and therapy response in patients with acute myeloid leukemia. Clin Exp Med 24, 57 (2024). https://doi.org/10.1007/s10238-023-01269-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10238-023-01269-w