Abstract
Objectives
Whole genome sequencing (WGS) is an emerging tool in clinical diagnostics. However, little has been said about its procedure costs, owing to a dearth of related cost studies. This study helps fill this research gap by analyzing the execution costs of WGS within the setting of German clinical practice.
Methodology
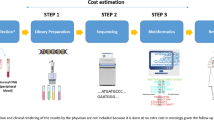
First, to estimate costs, a sequencing process related to clinical practice was undertaken. Once relevant resources were identified, a quantification and monetary evaluation was conducted using data and information from expert interviews with clinical geneticists, and personnel at private enterprises and hospitals. This study focuses on identifying the costs associated with the standard sequencing process, and the procedure costs for a single WGS were analyzed on the basis of two sequencing platforms—namely, HiSeq 2500 and HiSeq Xten, both by Illumina, Inc. In addition, sensitivity analyses were performed to assess the influence of various uses of sequencing platforms and various coverage values on a fixed-cost degression.
Results
In the base case scenario—which features 80 % utilization and 30-times coverage—the cost of a single WGS analysis with the HiSeq 2500 was estimated at €3858.06. The cost of sequencing materials was estimated at €2848.08; related personnel costs of €396.94 and acquisition/maintenance costs (€607.39) were also found. In comparison, the cost of sequencing that uses the latest technology (i.e., HiSeq Xten) was approximately 63 % cheaper, at €1411.20.
Conclusions
The estimated costs of WGS currently exceed the prediction of a ‘US$1000 per genome’, by more than a factor of 3.8. In particular, the material costs in themselves exceed this predicted cost.

Similar content being viewed by others
References
Bentley, D.R., et al.: Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456(7218), 53–59 (2008). doi:10.1038/nature07517
Rockman, M.V.: Reverse engineering the genotype-phenotype map with natural genetic variation. Nature 456(7223), 738–744 (2008). doi:10.1038/nature07633
Shendure, J., Ji, H.: Next-generation DNA sequencing. Nat. Biotechnol. 26(10), 1135–1145 (2008). doi:10.1038/nbt1486
Sanger, F., Nicklen, S., Coulson, A.R.: DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74(12), 5463–5467 (1977)
Renkema, K.Y., Stokman, M.F., Giles, R.H.: Knoers, Nine V A M: next-generation sequencing for research and diagnostics in kidney disease. Nat. Rev. Nephrol. 10(8), 433–444 (2014). doi:10.1038/nrneph.2014.95
Grada, A., Weinbrecht, K.: Next-generation sequencing: methodology and application. J. Invest. Dermatol. 133(8), e11 (2013). doi:10.1038/jid.2013.248
Buermans, H.P.J., den Dunnen, J.T.: Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta 1842(10), 1932–1941 (2014). doi:10.1016/j.bbadis.2014.06.015
Quail, M.A., Smith, M., Coupland, P., Otto, T.D., Harris, S.R., Connor, T.R., Bertoni, A., Swerdlow, H.P., Gu, Y.: A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 13, 341 (2012). doi:10.1186/1471-2164-13-341
Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., Lin, D., Lu, L., Law, M.: Comparison of next-generation sequencing systems. J Biomed. Biotechnol 2012, 251364 (2012). doi:10.1155/2012/251364
Green, E.D., Guyer, M.S.: Charting a course for genomic medicine from base pairs to bedside. Nature 470(7333), 204–213 (2011). doi:10.1038/nature09764
Kingsmore, S.F., Saunders, C.J.: Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Science translational medicine 3(87), 87ps23 (2011). doi:10.1126/scitranslmed.3002695
Choi, M., Scholl, U.I., Ji, W., Liu, T., Tikhonova, I.R., Zumbo, P., Nayir, A., Bakkaloğlu, A., Ozen, S., Sanjad, S., Nelson-Williams, C., Farhi, A., Mane, S., Lifton, R.P.: Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA 106(45), 19096–19101 (2009). doi:10.1073/pnas.0910672106
Carlson, C.S., Eberle, M.A., Kruglyak, L., Nickerson, D.A.: Mapping complex disease loci in whole-genome association studies. Nature 429(6990), 446–452 (2004). doi:10.1038/nature02623
Meuwissen, T., Goddard, M.: Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185(2), 623–631 (2010). doi:10.1534/genetics.110.116590
ACMG Board of Directors: Points to consider in the clinical application of genomic sequencing. Genetics Med Off J Am Coll Med Gen 14(8), 759–761 (2012). doi:10.1038/gim.2012.74
Sawyer, S.L., Hartley, T., Dyment, D.A., Beaulieu, C.L., Schwartzentruber, J., Smith, A., Bedford, H.M., Bernard, G., Bernier, F.P., Brais, B., Bulman, D.E., Warman Chardon, J., Chitayat, D., Deladoëy, J., Fernandez, B.A., Frosk, P., Geraghty, M.T., Gerull, B., Gibson, W., Gow, R.M., Graham, G.E., Green, J.S., Heon, E., Horvath, G., Innes, A.M., Jabado, N., Kim, R.H., Koenekoop, R.K., Khan, A., Lehmann, O.J., Mendoza-Londono, R., Michaud, J.L., Nikkel, S.M., Penney, L.S., Polychronakos, C., Richer, J., Rouleau, G.A., Samuels, M.E., Siu, V.M., Suchowersky, O., Tarnopolsky, M.A., Yoon, G., Zahir, F.R., Majewski, J., Boycott, K.M.: Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin. Gen. (2015). doi:10.1111/cge.12654
Berg, J.S., Khoury, M.J., Evans, J.P.: Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genetics Med. 6, 499–504 (2011). doi:10.1097/GIM.0b013e318220aaba
Ford, D., Easton, D.F., Stratton, M., Narod, S., Goldgar, D., Devilee, P., Bishop, D.T., Weber, B., Lenoir, G., Chang-Claude, J., Sobol, H., Teare, M.D., Struewing, J., Arason, A., Scherneck, S., Peto, J., Rebbeck, T.R., Tonin, P., Neuhausen, S., Barkardottir, R., Eyfjord, J., Lynch, H., Ponder, B., Gayther, S.A., Birch, J.M., Lindblom, A., Stoppa-Lyonnet, D., Bignon, Y., Borg, A., Hamann, U., Haites, N., Scott, R.J., Maugard, C.M., Vasen, H., Seitz, S., Cannon-Albright, L.A., Schofield, A., Zelada-Hedman, M.: Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am. J. Human Gen. 62(3), 676–689 (1998). doi:10.1086/301749
Green, R.C., Berg, J.S., Grody, W.W., Kalia, S.S., Korf, B.R., Martin, C.L., McGuire, A.L., Nussbaum, R.L., O’Daniel, J.M., Ormond, K.E., Rehm, H.L., Watson, M.S., Williams, M.S., Biesecker, L.G.: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Gen. Med. Off. J. Am. Coll. Med. Gen. 15(7), 565–574 (2013). doi:10.1038/gim.2013.73
Knoppers, B.M., Deschênes, M., Zawati, M.H., Tassé, A.M.: Population studies: return of research results and incidental findings policy statement. Eur. J. Human Gen. EJHG 21(3), 245–247 (2013). doi:10.1038/ejhg.2012.152
Evans, J.P., Skrzynia, C., Burke, W.: The complexities of predictive genetic testing. BMJ 322(7293), 1052–1056 (2001)
Dondorp, W.J., de Wert, W.J., Guido, M.W.R.: The ‘thousand-dollar genome’: an ethical exploration. Eur. J. Hum. Genet. 21(Suppl 1), S6–26 (2013). doi:10.1038/ejhg.2013.73
McBride, C.M., Koehly, L.M., Sanderson, S.C., Kaphingst, K.A.: The behavioral response to personalized genetic information: will genetic risk profiles motivate individuals and families to choose more healthful behaviors? Annu. Rev. Public Health 31, 89–103 (2010). doi:10.1146/annurev.publhealth.012809.103532
Ten Bosch, John, R., Grody, W.W.: Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. J Mol. Diagn JMD 10(6), 484–492 (2008). doi:10.2353/jmoldx.2008.080027
National Human Genome Research Institute: The Human Genome Project Completion: Frequently Asked Questions (2010). https://www.genome.gov/11006943. Assessed 17 July 2015
Wetterstrand, K.A.: DNA sequencing costs: data from the NHGRI Genome Sequencing Program. http://www.genome.gov/sequencingcosts. Assessed 13 January 2015
Collins, F.: Has the revolution arrived? Nature 464(7289), 674–675 (2010). doi:10.1038/464674a
Hayden, E.C.: Technology: the $1,000 genome nature. Nature 507(7492), 294–295 (2014). doi:10.1038/507294a
Service, R.F.: Gene sequencing. The race for the $1000 genome. Science (New York, N.Y.) 311(5767), 1544–1546 (2006). doi:10.1126/science.311.5767.1544
Frank, M., Prenzler, A., Eils, R., Graf von der Schulenburg, J-M.: Genome sequencing: a systematic review of health economic evidence. Health economics review 3(1), 29 (2013). doi: 10.1186/2191-1991-3-29
Götze, U., Northcott, D., Schuster, P.: Investment appraisal. Methods and models. Springer texts in business and economics
Illumina, Inc.: cBot (2015). http://www.illumina.com/products/cbot.html. Assessed 17 March 2015
Holt, R.A., Jones, Steven J M: The new paradigm of flow cell sequencing. Genome research 18(6), 839–846 (2008). doi:10.1101/gr.073262.107
QIAGEN: QIAamp DNA Blood Mini Kit (2016). https://www.qiagen.com/de/shop/sample-technologies/dna/dna-preparation/qiaamp-dna-blood-mini-kit#orderinginformation. Assessed 01 March 2016
Meynert, A.M., Ansari, M., FitzPatrick, D.R., Taylor, M.S.: Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinfo. 15, 247 (2014). doi:10.1186/1471-2105-15-247
Dewey, F.E., Pan, S., Wheeler, M.T., Quake, S.R., Ashley, E.A.: DNA sequencing: clinical applications of new DNA sequencing technologies. Circulation 125(7), 931–944 (2012). doi:10.1161/CIRCULATIONAHA.110.972828
Schlötterer, C., Tobler, R., Kofler, R., Nolte, V.: Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 15(11), 749–763 (2014). doi:10.1038/nrg3803
Kofler, R., Schlötterer, C.: A guide for the design of evolve and resequencing studies. Mol. Biol. Evol. 31(2), 474–483 (2014). doi:10.1093/molbev/mst221
McCourt, C.M., McArt, D.G., Mills, K., Catherwood, M.A., Maxwell, P., Waugh, D.J., Hamilton, P., O’Sullivan, J.M., Salto-Tellez, M.: Validation of next generation sequencing technologies in comparison to current diagnostic gold standards for BRAF, EGFR and KRAS mutational analysis. PLoS One 8(7), e69604 (2013). doi:10.1371/journal.pone.0069604
Karow, J.: In Sequence 2013 Survey: Illumina Pulls Further Ahead, Interest in Oxford Nanopore Remains High (2014)
Studt, T,: Innovations Drive Rapid NGS Growth. Lower costs, high throughput and enhanced accuracies provide next-generation sequencing users with enhanced medical information. (2015)
PHG Foundation: PHG Foundation: Next steps in the sequence. The implications of whole genome sequencing for health in the UK, Cambridge (2011). http://www.phgfoundation.org/file/10363/. Assessed 23 April 2015
National Human Genome Research Institute: Genetic Discrimination (2014). http://www.genome.gov/10002077. Accessed 23 April 2015
American Society of Clinical Oncology policy statement update: Genetic testing for cancer susceptibility. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 21(12), 2397–2406 (2003). doi:10.1200/JCO.2003.03.189
Statement of the ESHG on direct-to-consumer genetic testing for health-related purposes. European journal of human genetics: EJHG 18(12), 1271–1273 (2010). doi:10.1038/ejhg.2010.129
Acknowledgments
We thank the German Cancer Research Center (DKFZ), Heidelberg; Illumina Inc., San Diego, CA, USA; Hanover Medical School (MHH); German Faculty of Mathematics and Computer Science Chair of RNA Bioinformatics and High Throughput Analysis at the Friedrich-Schiller-University, Jena; Max Planck Institute, Berlin; Center for Human Bioinformatics, Heidelberg; Fraunhofer Institute for Interfacial Engineering and Biotechnology IGB, Stuttgart; Bernhard Nocht Institute for Tropical Medicine, Hamburg; Helmholtz Centre, Munich; Bioinformatics and System Biology Justus Department at the Liebig-University Gießen; and the Institute for Biochemistry, Biotechnology and Bioinformatics at the Technical University of Braunschweig and BioNTech RNA Pharmaceuticals GmbH for their valuable information.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Plöthner, M., Frank, M. & von der Schulenburg, JM.G. Cost analysis of whole genome sequencing in German clinical practice. Eur J Health Econ 18, 623–633 (2017). https://doi.org/10.1007/s10198-016-0815-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10198-016-0815-0