Abstract
The metabotropic glutamate receptors (mGluRs) are a class of G-protein-coupled receptor that undergo extensive interactions with scaffolding proteins, and this is intrinsic to their function as an important group of neuromodulators at glutamatergic synapses. The Caenorhabditis elegans nervous system expresses three metabotropic glutamate receptors, MGL-1, MGL-2 and MGL-3. Relatively little is known about how the function and signalling of these receptors is organised in C. elegans. To identify proteins that scaffold the MGL-1 receptor, we have conducted a yeast two-hybrid screen. Three of the interacting proteins, MPZ-1, NRFL-1 and PTP-1, displayed motifs characteristic of mammalian mGluR scaffolding proteins. Using cellular co-expression criterion, we show mpz-1 and ptp-1 exhibited overlapping expression patterns with subsets of mgl-1 neurons. This included neurones in the pharyngeal nervous system that control the feeding organ of the worm. The mGluR agonist L-CCG-I inhibits the activity of this network in wild-type worms, in an MGL-1 and dose-dependent manner. We utilised L-CCG-I to identify if MGL-1 function was disrupted in mutants with deletions in the mpz-1 gene. The mpz-1 mutants displayed a largely wild-type response to L-CCG-I, suggesting MGL-1 signalling is not overtly disrupted consistent with a non-obligatory modulatory function in receptor scaffolding. The selectivity of the protein interactions and overlapping expression identified here warrant further investigation of the functional significance of scaffolding of metabotropic glutamate receptor function.
Similar content being viewed by others
Introduction
Glutamate is the major excitatory neurotransmitter of the mammalian central nervous system, and it signals through two classes of receptor, ionotropic receptors (ligand-gated ion channels) and metabotropic glutamate receptors (G-protein-coupled receptors). There are eight mammalian metabotropic glutamate receptors that are divided into three subgroups (Group I-III) based upon sequence homology, pharmacology and signalling (Conn and Pin 1997). The mGluRs are non-essential, but perform pivotal modulatory roles in health and disease (Riedel et al. 1996; Riedel and Reymann 1996). The mGluR subtypes are specifically targeted either pre- or post-synaptically, and they are localised to discrete subcellular domains. Group I receptors (mGluR1/5) tend to be post-synaptic and localised to the peri-synaptic compartment (Luján et al. 1997; Nusser et al. 1994), Group II receptors (mGluR2/3) can be localised pre-and post-synaptically and are localised in pre-terminal membranes (Lujan et al. 1996; Azkue et al. 2000) and Group III receptors (for example mGluR7) tend to be localised pre-synaptically, close to the sight of neurotransmitter release (Shigemoto et al. 1996; Blümcke et al. 1996). In addition, it has been shown that mGluRs can engage in signalling crosstalk with plasma membrane ion channels, in a manner that is independent of G-protein signalling (Perroy et al. 2001).
The mGluRs have distinct domain architecture, consisting of a large extracellular N-terminal domain containing the agonist binding site and a cysteine-rich region, intracellular loops that direct G-protein interactions and an intracellular C-terminal domain (Conn and Pin 1997). The targeting, discrete sublocalisation and the signalling cascades of mGluRs can be specified by protein–protein interactions between intracellular proteins and the intracellular C-terminal domain of the receptor. A number of these intracellular proteins have been identified as interacting with specific motifs of the receptor C-terminal and are described as scaffolding proteins. Perhaps, the most conserved motif across the mGluR family is the PDZ motif and a number of proteins that perform roles in the scaffolding of receptor function interact via a PDZ-binding domain (Perroy et al. 2001). Alternative splicing of the C-terminal can create isoforms containing different combinations of scaffolding motifs, generating further functional diversity amongst the mGluR family (Perroy et al. 2001). A number of proteins that interact with the intracellular C-terminal of the mammalian receptors were identified using biochemical protein-interaction approaches, which have included the yeast two-hybrid system (Perroy et al. 2001).
The Caenorhabditis elegans genome encodes three metabotropic glutamate receptors, named MGL-1, MGL-2 and MGL-3. Sequence analysis suggests the three receptors each representing orthologues of one of the mammalian mGluR subfamilies, and the functional diversity within the mammalian family is conserved in C. elegans (Dillon et al. 2015). There is currently a limited understanding of this class of receptor, compared to the ionotropic families of glutamate receptors (viz NMDA, AMPA and Ivermectin channels) in C. elegans. We have previously shown that the C. elegans mGluR orthologue, MGL-1, is expressed in the pharyngeal nervous system. We have probed its function using the mGluR agonist L-CCG-I to show that it is a negative regulator of pharyngeal network activity. MGL-1 is required for coordinating plasticity in feeding behaviour and reproduction upon the removal of food (Dillon et al. 2015; Jeong and Paik 2017). To build upon this, we have identified candidate MGL scaffolding proteins and screened for their potential to modulate the activity of the pharyngeal network. We performed a yeast two-hybrid screen using the C-terminal of the MGL-1 receptor and used it to define potential selectivity of three candidate scaffolding partners. The molecules PTP-1, NRFL-1 and MPZ-1 contain PDZ-binding domains that robustly bound to the C-terminal domain of the receptor in the yeast two-hybrid system. We characterised these candidates and showed that PTP-1 did not selectively bind to MGL-1. NRFL-1 and MPZ-1 did bind selectively to MGL-1, and other data have highlighted that MPZ-1 also binds to targets outside of the panel of targets used in this study (Xiao et al. 2006). Our subsequent expression studies show that ptp-1 and mpz-1 but not nrfl-1 are co-expressed with mgl-1 in discrete subsets of neurones. Overall, our data identify the potential for the scaffolding of MGL-1 receptor function, which will provide additional routes to characterise the function of MGL-1 in C. elegans and provide a model to investigate dysfunction and function associated with this class of receptor.
Methods
Genetics
All C. elegans strains were cultured at 20 °C under conditions previously described (Brenner 1974). Bristol N2 worms were used as the wild-type strain. Mutant alleles used in this study were as follows: mgl-1(tm1181), mpz-1(tm1136) and the strain VC559 carrying the allele mpz-1(gk273)/mIn1[dpy-10(e128) mIs14(myo-2::GFP)]. The strain carrying allele mgl-1(tm1811) and the strain VC559 were both outcrossed three and one times, respectively. The strain mpz-1(tm1136/gk273) was generated by mating mpz-1(tm1136) males with mpz-1(gk273)/mIn1[dpy-10(e128) mIs14(myo-2::GFP)] hermaphrodites. mpz-1(tm1136/gk273) hermaphrodites were selected in the F1 generation by the absence of pharyngeal GFP (marking the mIn1 inversion). The integrated strain utIs35(mgl-1::GFP) was kindly provided by Professor Katsura [previously described (Dillon et al. 2015)]. The strain pha-1(e2123) was kindly provided by R Schnabbel and used to generate the mgl-1::GFP;pha-1(e2123) selectable marker strain for the co-expression analysis of yeast two-hybrid candidates.
Yeast two-hybrid screen
The intracellular C-terminal domain of MGL-1, NMR-1, GLR-1 and LET-23 was amplified by PCR from a C. elegans cDNA (Origene Technologies Inc.). PCR products were subcloned EcoRI-SalI into pGilda containing the LexA DNA-binding domain. To test for nuclear targeting and expression, the bait plasmid was transformed into EGY48 yeast cells with the plasmid pJK101, which contains 2xOlexA-lacZ downstream of the inducible GAL-1 promoter. Transformants were plated onto GAL-H-U plates containing X-GAL (80 mg/L), and white colonies after 3 days at 30 °C indicated BD-MGL-1 was being nuclear targeted and expressed due to the suppression of lacZ expression. To test for the auto-activation of the 8xOlexA-lacZ reporter gene, each of the bait plasmids was co-transformed into EGY48 cells with the plasmid pSH18-34 (8xOlexA-lacZ reporter), plated onto GAL-His-Ura + X-GAL media and placed at 30 °C. Blue colour after 20 days indicated auto-activation. To test for auto-activation of 6xOlexA-LEU2 reporter genes, each of the bait plasmids was co-transformed into EGY48 cells with pSH18-34 and pJG4-5 (the empty prey vector), plated onto GAL–His–Ura–Trp–Leu media. Growth after 20 days at 30 °C indicated auto-activation of the LEU2 reporter gene by the bait in the absence of a prey clone.
After confirming the MGL-1 bait construct was nuclear targeted, expressed and did not auto-activate the reporter genes, we proceeded to perform a yeast two-hybrid screen to identify MGL-1 interacting proteins. The screen was performed with the DupLex-A™ yeast two-hybrid system (Origene). The cDNA library was constructed using an oligo d(T) primer and cloned into the EcoRI-XhoI sites of the prey vector pJG4-5. EGY48 cells were transformed with the plasmids pSH18-34 and selected for on GLU–Ura media; these cells were then transformed with the MGL-1 C-terminal bait (BD-MGL-1) and selected for on GLU–His–Ura media. These cells were then transformed with the C. elegans cDNA library pJG4-5 and selected on GLU–Trp–His–Ura. The number of transformants screened was titred to 400 × 106 corresponding to approximately three times the titre of viable transformants. The transformants were plated onto media containing galactose (GAL) and –Trp–His–Ura–Leu at a density of 2 × 106/plate and incubated at 30 °C. On days 4, 5 and 7, post-plating colonies were transferred to GAL–Trp–His–Ura–Leu + X-Gal. On day 10, β-galactosidase filter lift assays were performed (as described in (Breeden and Nasmyth 1985)) on the screening plates, and colonies positive for blue colour were identified and transferred onto media GAL–Trp–His–Ura–Leu + X-Gal. Yeast colonies that grew on GAL–Trp–His–Ura–Leu + X-Gal and were blue in colour after 5 days at 30 °C were re-streaked onto GAL–Trp–His–Ura–Leu + X-Gal. Those colonies that grew on GAL–Trp–His–Ura–Leu + X-Gal and were blue in colour after 4 rounds of selection were further characterised.
Following this, the plasmids from individual transformants were extracted and transformed into bacteria followed by selection. Hae 1 was used to profile the variation in the number of clones, and representative clones from each of the prey plasmids were sequenced from the 5′ end to define the transactivation domain which was in-frame with the candidate interacting protein. This analysis involved screening through 179 potential clones, and after identifying cDNA, we iterated authenticity on the basis of in-frame, existing as multiple transformants and preferably existing representative clones. Using this approach, the number of original clones was pared down to 21 by excluding out-of-frame clones and those associated with nuclear function (Online Resource 1). Three candidates were identified by more than one independent clone. This is assimilated in Table 1.
Molecular biology
The 5′RACE products were obtained from two rounds of amplification using a 5′RACE primer in the first reaction and a nested 5′RACE reaction in the second reaction, which was performed using a nested 5′RACE primer and either an SL-1 primer or RACE adaptor primer. cDNA for RACE analysis was synthesised using the SMART RACE cDNA amplification kit from Clontech™. 1 µg of total RNA extracted from a mixed stage population of worms was used, and the RACE adaptor fused first-strand cDNA served as the template for the RACE PCR. The RACE primers were designed with the available sequence information from Wormbase (WS253). The RACE products were gel-purified and TOPO-cloned (Invitrogen) and then sequenced. Isoforms are named according to the annotation in Wormbase WS253.
Promoter fusion constructs were generated from genomic DNA fragments cloned into the vector backbone pHAB (provided by Dr Howard Baylis) and GFP replaced by either dsRED2 or mRFP-1. The mpz-1::mrfp-1 construct included 2315 bp of 5′UTR, the nrfl-1::dsRed2 construct included 3258 bp of 5′UTR and the ptp-1::mRFP-1 construct contained 2652 bp of 5′UTR. mRFP-1 was obtained from Dr. M. Hannah. The promoter constructs were microinjected into the selectable marker strain mgl-1::GFP;pha-1(e2123) at 30 ng/μl with the transformation plasmid marker pha-1 at 50 ng/μl. The transformants were selected by their growth at 25 °C, and three transgenic lines were analysed for co-expression.
For the mpz-1 transcript analysis, total RNA was extracted from a mixed stage population of worms and 5 μg was used to perform the first-strand cDNA synthesis (Superscript II, Invitrogen). The first-strand cDNA synthesis was performed using an mpz-1 gene-specific primer, and 2 μl of the first-strand cDNA synthesis mix was used in the PCR reaction.
Microscopy
Worms were mounted on 2% agarose pads and immobilised for imaging using 20 mM Na-Azide. mpz-1::mRFP-1;mgl-1::GFP confocal images were recorded using Zeiss LSM510 laser scanning confocal microscope. The settings were Objective 63 × Oil DIC, GFP excitation 488 nm, band pass filter 505–530 nm, mRFP-1 excitation 543 nm, band pass filter 560–615 nm. Optical sections were collected with a step size of 0.98 μM. ptp-1::mRFP-1;mgl-1::GFP, and nrfl-1::mRFP-1 images were captured using a Hamamatsu ORCA camera attached to a Nikon Eclipse E800 microscope, Objective × 60 Oil DIC and × 20 DIC, respectively. Images were analysed using ImageJ (NIH).
Electrophysiological recordings
Electropharyngeograms were performed on L4 plus two- or three-day-old adult worms. The recordings were made from a semi-intact pharyngeal preparation as previously described (Dillon et al. 2015) in a modified Dent’s saline solution (144 mM NaCl, 10 mM MgCl2, 1 mM CaCl2, 6 mM KCl, 10 mM Glucose and 5 mM HEPES; pH 7.4). L-CCG-I dose response curves were generated by adding increasing concentrations to individual worms, with a 5 min wash period between each application of the drug. The wash period immediately prior to drug application was used to calculate the percentage change in the rate of pumping. L-CCG-I was supplied by TOCRIS Biosciences and diluted to the required concentration in Dent’s saline. During pharmacological experiments, solutions were added to and removed from the recording chamber using a 1 ml pipette. Dose response curves were plotted using GraphPad Prism 7, and the EC50 for each strain was calculated. The variation in the dose response curves was investigated using a two-way ANOVA test.
Results
Yeast two-hybrid screen
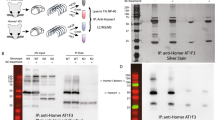
The intracellular C-terminal of MGL-1 was fused in-frame to the LexA DNA-binding domain (BD) protein in the vector pGilda, to create the bait plasmid construct (BD-MGL-1). The amino acid sequence of the intracellular C-terminal used is shown in Fig. 1, and it contains a predicted Type I PDZ-binding motif with the consensus sequence S/T-X-Φ, where Φ denotes a hydrophobic amino acid (usually V/I/L). We confirmed the MGL-1 C-terminal bait was expressed, nuclear targeted and that it did not auto-activate the expression of either of the reporter genes, LEU2 and lacZ (Fig. 1). The BD-MGL-1 bait was co-transformed into the yeast strain EGY48 with pSH18-34 (a plasmid containing 8xOlexA-lacZ reporter) and the C. elegans DupLex-A yeast two-hybrid system prey library (Origene). The prey library was prepared using mRNA from a mixed stage population of wild-type worms. The number of transformants screened was titrated and plated onto –HTUL(GAL) media to select for colonies expressing the LEU2 reporter gene. Colonies growing were selected up to 10 days post-plating and replica plated onto –HTUL(GAL) plates containing X-GAL to select for transformants expressing the reporter gene lacZ, encoding β-Galactosidase as well as LEU2 (Fig. 1). Colonies that exhibited a strong expression of both of the reporter genes after four rounds of selection were prioritised for further analysis. Independent clones identified three interactions: NRFL-1, PTP-1 and MPZ-1 (Table 1). These three candidates were indicated through independent clones encoding the candidate proteins PDZ domains. These were prioritised and further analysis showed that the longest identified clones of NRFL-1, PTP-1 and MPZ-1 did not auto-activate either of the reporter genes (Fig. 2). NRFL-1 corresponds to the isoform C01F6.6a and contains two tandem PDZ-binding domains; it is predicted to be cytoplasmic, and it is the single orthologue of the mammalian NHERF (Na+/H+ exchange regulatory factor) family of genes (Hagiwara et al. 2012). PTP-1 contains an N-terminal FERM (4.1 protein, Ezrin, Radixin and Moesin) domain, known to direct cytoskeletal interactions, followed by a PDZ domain and a C-terminal tyrosine phosphatase domain. Each of the PTP-1 identified clones contains a region that is common to each of the predicted PTP-1 isoforms (C48D5.2a-c), but does not contain the N-terminal FERM domain of the longest variant (C48D5.2a, referred to as PTP-1a in this study). PTP-1 is an orthologue of the mammalian gene PTPMEG. MPZ-1 is an orthologue of the mammalian multi-PDZ protein MUPP-1 (Xiao et al. 2006). MPZ-1 is a multi-PDZ domain protein; the longest MPZ-1 isoform C52A11.4d contains 10 PDZ domains. The longest MPZ-1 prey clone identified corresponds to the 3′ region of MPZ-1 and contains four PDZ domains.
Yeast two-hybrid screen with the C. elegans MGL-1 receptor C-terminal domain. a MGL-1 C-terminal used as the bait, BD-MGL-1CT. The predicted Type I PDZ motif is highlighted in bold and underlined. b Yeast two-hybrid control assays confirmed the BD-MGL-1 bait construct is nuclear targeted and expressed (colonies appear white) and it does not auto-activate either of the reporter genes LEU2 (no growth in the absence of Leucine) and lacZ (colonies appear white). c Transformants identified from the screening plates were taken through four rounds of selection on –HTUL(Gal) + X-Gal plates. Transformants that expressed reporter genes, LEU2 and lacZ, after the four rounds of selection were further characterised
Analysis of candidate interactors. a The domain architecture of the three prioritised proteins, MPZ-1 (C52A11.4a), NRFL-1(C01F6.2a) and PTP-1(C48D5.2a). The dashed lines indicate the region contained within the longest prey clone identified and used for subsequent analysis. Domains are coded as PDZ domain: Grey, FERM domain: Red, Tyrosine Phosphatase: Green and L27 domain: Orange. Scale bar: 100 amino acids. b Prey clones were each transformed into EGY48 cells and plated onto –TL(GAL) media. In the absence of Leucine, the cells did not grow confirming the prey clones did not auto-activate LEU2. Prey clones were co-transformed with the plasmid pSH18-34, which carries the lacZ reporter gene and plated onto –TU(GAL) + X-GAL. Streaked patches remained white, confirming the prey clones did not auto-activate lacZ. Images were recorded after 10 days of incubation at 30 °C. c The prey clones prioritised from the MGL-1 two hybrid screen were screened against the intracellular C-terminal of the receptors GLR-1, NMR-1 and LET-23. BD-MGL-1 was included as a control. The C-terminal of NMR-1 contains a Type II PDZ consensus motif, defined as Φ-X-Φ, where Φ denotes a hydrophobic residue. An interaction between GLR-1 and PTP-1 was identified, indicated by the blue colouration and growth on media –HUTL(GAL) + X-Gal. Images were taken after 3 days at 30 °C (colour figure online)
Probing the specificity of the interaction between the MGL-1 C-terminal and MPZ-1, NRFL-1 and PTP-1
To assess the selectivity of the three prey clones that exhibit the in vitro interaction with the intracellular C-terminal domain of MGL-1, we compared their interaction with NMR-1, GLR-1 and LET-23. Each has a C-terminal PDZ-binding motif, and it has previously been shown that the intracellular C-terminal is important in directing the function of the receptor. The intracellular C-terminal of the ionotropic glutamate receptor subunits NMR-1 and GLR-1 and the tyrosine kinase receptor LET-23 was fused in-frame with the LexA-binding domain to create the bait plasmid constructs BD-NMR-1, BD-GLR-1 and BD-LET-23. We confirmed that each of the bait constructs was being expressed, nuclear targeted and did not auto-activate either of the reporter genes. NRFL-1 and MPZ-1 prey clones did not interact with any of the C-terminal bait constructs, and PTP-1 interacted only with the C-terminal of the GLR-1 receptor (Fig. 2). This in vitro interaction of PTP-1 with MGL-1 and GLR-1 indicates that it may perform a role in scaffolding distinct classes of glutamate receptor.
5′ RACE characterisation of mpz-1, nrfl-1 and ptp-1 and analysis of co-expression with mgl-1
To identify if the MGL-1 interaction predicted potential functional significance, we investigated the cellular expression, of mgl-1 and the identified PDZ domain containing proteins, in the worm. To do this, we designed fluorescent reporter constructs reporting on the cellular expression of each interacting gene. Our approach was based on taking extended regions of the 5′ end of the gene encompassing at least 2.3 Kb upstream of the predicted start. The first stage of this approach required the characterisation of the 5′ end of each of the genes as this region of these genes is poorly annotated in the database and we found it revealed important elements of the organisation of other genes (Dillon et al. 2015). To assist in this, we performed a 5′ RACE (Fig. 3). Where the 5′RACE reaction produced multiple bands, a nested 5′ RACE was performed on the first-round reaction. The amplified products were cloned and then sequenced. The longest NRFL-1 nested 5′ RACE product cloned and sequenced was SL-1 trans-spliced; it contained the first 2 exons, and it confirmed the predicted ATG start codon defined by the gene model annotated as C01F6.6a in Wormbase. In contrast, the PTP-1 5′ RACE reaction produced multiple bands that were not of the predicted size. A nested 5′ RACE reaction yielded three prominent amplicons. The longest highlighted the previously predicted start codon and suggests a 42 bp 5′ UTR. It confirmed the intron/exon boundaries described by the C48D5.2a gene model for exons 1, 2 and 3. RACE of MPZ-1 was performed using an SL-1 primer in combination with a nested mpz-1 primer and identified three different SL-1 trans-spliced isoforms. Two of the three SL-1 trans-spliced isoforms confirmed the transcripts annotated as mpz-1a/d and mpz-1b in (Xiao et al. 2006). The third trans-spliced isoform defines an exon upstream of the first exon defined by mpz-1a/1d in (Xiao et al. 2006) and confirms the 5′ end of the gene model annotated as C52A11.4a in Wormbase.
5′ RACE characterisation of MPZ-1, PTP-1 and NRFL-1. The analysis was performed on RACE cDNA prepared from total RNA extracted from a mixed stage population of wild-type worms. Exons contained within RACE products and confirmed experimentally are highlighted in black. The location of the primers used to perform the RACE PCR is indicated (P′). The position of SL-1 trans-splicing is indicated by ‘S’. Scale bars = 1000 bp (colour figure online)
Using the information obtained from the 5′RACE analysis, we made reporter constructs such that at least 2.3 Kb 5′ of the putative start codon was cloned upstream of dsRed2 or mRFP-1. These constructs report on the cellular expression pattern of mpz-1, nrfl-1 and ptp-1 (see Fig. 4). This approach maximises the ability to generate constructs with good authenticity of the physiological expression, although we cannot rule out important roles for downstream introns in modulating expression patterns. The constructs that drive expression of the fluorescent protein are not expected to harbour any targeting information and constructs cause expression of relatively soluble fusion proteins that fill the expressing cell. These constructs were injected into a pre-existing integrated line of worms expressing a translational fusion in which GFP is integrated into the C-terminal domain of MGL-1. As reported, this displays the cellular expression of mgl-1 (Dillon et al. 2015).
Co-expression of the two hybrid identified genes with mgl-1 in vivo. a Genomic regions of ptp-1(C48D5.2a), mpz-1(C52A11.4a) and nrfl-1(C01F6.6a) that were used to generate reporter constructs are shown. Scale bars = 500 bp b and c co-expression of mgl-1 with ptp-1 and mpz-1, respectively, in the pharyngeal nervous system (desheathed pharynx). In (C), co-expression in NSM is indicated by white arrowheads and co-expression in M4 is indicated by the blue arrowhead. In (C), sections XZ1-4 have been taken from the Z stack projection and show neuronal co-expression of mgl-1 and mpz-1 in the pharynx (arrowheads). Although co-expression was observed in additional regions, only those structures with an identified co-expression with mgl-1::GFP in the pharyngeal nervous system are shown. d Expression of nrfl-1::mRFP-1. Asterisks indicate the expression of nrfl-1 in the pharyngeal corpus muscle and the intestine. nrfl-1 was not co-expressed with mgl-1::GFP. The pattern of reporter expression was confirmed in at least two independent lines for each candidate interactor. Scale bars = 20 μM (colour figure online)
Analysis of the desheathed pharynx identified co-expression of mgl-1::GFP and ptp-1::mRFP-1 in the pharyngeal nervous system (Fig. 4b). mpz-1 was identified as being expressed in the pharyngeal neurones (Fig. 4c) (including M4 and NSM based on their size and relative position), extrapharyngeal neurones, body wall muscle and vulval muscle. Co-expression of mpz-1::mRFP-1 with mgl-1::GFP was identified in neurones of the nerve ring and the pharyngeal nervous system in the desheathed pharynx preparation (Fig. 4c). Hence, in the case of ptp-1 and mpz-1, widespread expression was identified, overlapping expression with mgl-1 was only identified in a subpopulation of these cells and mgl-1 was expressed in cells that did not express mpz-1 nor ptp-1. This suggests that MPZ-1 and PTP-1 are not obligatory for MGL-1 receptor function but well placed to interact with the receptor in some of the circuits in which it might modulate function.
We identified nrfl-1 expression in the intestine and pharyngeal muscle (Fig. 4d). This cellular expression is inconsistent with the specific neuronal expression of mgl-1. Furthermore, despite some expression in neurones in the pharynx and the tail, it was not co-expressed in any of the same neurones as mgl-1::GFP. Such a mismatch in expression would argue against a functional role for NRFL-1 scaffolding of MGL-1.
Electrophysiological analysis of MGL-1 interaction with MPZ-1
MGL-1 and the other metabotropic receptor mutants are viable, but exhibit discernible behavioural phenotypes (unpublished observations). In the case of MGL-1, we defined a modulatory role in C. elegans feeding behaviour that makes the pharynx sensitive to metabotropic glutamate agonist L-CCG-I (Dillon et al. 2015). In view of this and the identification of mpz-1 and mgl-1 co-expression in neurones belonging to the pharyngeal nervous system, we used L-CCG-I to assay for a modulatory role of mpz-1 in this circuit. Wormbase denotes two mpz-1 mutant alleles mpz-1(tm1136) and mpz-1(gk273)/mIn1[dpy-10(e128) mIs14(myo-2::GFP)] (Fig. 5). The allele mpz-1(tm1136) is a 1124 bp genomic deletion, with a 3 bp insertion. PCR amplification over the deletion region was performed on cDNA prepared from mpz-1(tm1136) mutant worms and identified a single band of the predicted size for the mutant, suggesting that this transcript is stable and does not undergo degradation (Fig. 5). Sequencing of the PCR fragment amplified identified a 167 bp deletion, within the 5′ end of the transcript corresponding to exons 9 and 10 of the isoform C52A11.4a annotated on Wormbase. The deletion is predicted to introduce a frame shift and a premature stop codon into each of the three mpz-1 SL-1 trans-spliced isoforms identified within this study. It is predicted that this mutant transcript will not generate a full length, fully functional protein.
Transcript analysis of the mpz-1 mutant strains mpz-1(gk273/+) and mpz-1(tm1136). A The isoform C52A11.4a is shown, with the location of SL-1 trans-splice sites (‘S’) identified in this study and putative start codons (‘M’) indicated. Primers were designed to amplify a short region containing the deletions defined by alleles gk273 and tm1136. (The region over which the PCR amplification was performed is indicated.) b PCR was performed on cDNA synthesised from total RNA prepared from a mixed stage population of N2, gk273/+ and tm1136 worms. PCR products of the predicted size were amplified from wild-type, gk273/+and tm1136 cDNA. The shorter amplification product corresponding to gk273 appears less intense on the gel than the wild-type product, suggesting this transcript is less abundant in the gk273/+ worms. The band amplified corresponding to tm1136 appears as intense as the wild-type control, suggesting this transcript has a similar abundance in the tm1136 worms. 5 μl samples were removed from the PCR reaction after 25 and 32 cycles for DNA gel electrophoresis. For each strain tested n = 2, representative images are shown
Interestingly, the gk273 allele is homozygous lethal, and it is a 1183 bp genomic deletion and 8 bp insertion. The strain VC559 used in this study has the genotype mpz-1(gk273)/mIn1[dpy-10(e128) mIs14(myo-2::GFP)]; hence, it carries the gk273 allele, and it is chromosome balanced by the GFP and dpy-10 marked inversion mIn1. For this reason, it is heterozygous for the gk273 deletion allele. The same transcript analysis described for the tm1136 allele was performed on cDNA generated from mpz-1(gk273)/mIn1[dpy-10(e128) mIs14(myo-2::GFP)] worms to characterise the effect of the gk273. PCR amplification yielded two bands, corresponding to the wild type and the mutant transcript (Fig. 5). Sequencing of the mutant fragment amplified identified a 152 bp deletion at the 5′ end of mpz-1, corresponding to exon 6 of the isoform C52A11.4a. The deletion is predicted to introduce a premature stop codon into only one of the mpz-1 SL-1 trans-spliced variants identified in our study. In the case of the other two SL-1 trans-splice variants, the deletion removes an untranslated exon that resides within the 5′UTR and upstream of the predicted start codon defined by these shorter isoforms. Thus, the more profound consequence of this genomic disruption is unclear, but analysis of the phenotypic segregation of mpz-1(gk273/+) progeny identified gk273 homozygotes has an early developmental phenotype in which embryos arrest 3–6 h after being laid, corresponding to early gastrulation (data not shown).
All of the mpz-1 mutants appeared normal in terms of the basal rate of pharyngeal pumping. Worms homozygous for the allele mpz-1(tm1136) displayed a wild-type response to L-CCG-I (see Fig. 6) in that the response of the pharynx to the mGluR agonist was a profound inhibition of pharyngeal pumping. Furthermore, the efficacy of the inhibition was similar as the EC50 for inhibition was similar to both wild-type and mutant stains. The strain VC559 carrying the mpz-1(gk273) deletion is heterozygous, mpz-1(gk273/+) and displayed a wild-type response to L-CCG-I (see Fig. 6). Further, the strain carrying the allele mpz-1(tm1136) was crossed with worms carrying the mpz-1(gk273) allele to generate mpz-1(tm1136/gk273) worms. These also displayed a wild-type response to L-CGG-I and similar to that of mpz-1(tm1136) and mpz-1(gk273/+) worms.
L-CCG-I responses of the three mpz-1 mutant strains. a Electropharyngeogram recordings were made from wild-type (N2) and each of the mpz-1 mutants across a range of L-CCG-1 doses. Drug application is indicated by black bars, and the dose is stated beneath. b L-CCG-I dose–response plot for mpz-1 mutants and wild type. The mpz-1 mutants were inhibited by L-CCG-I at 3 mM and 10 mM, a profile that is akin to the N2 wild-type strain. The EC50 was calculated as gk273/+ 2.68 μM; tm1136 2.92 μM; gk273/tm1136 1.1 μM; wild-type 2.47 μM. Each point represents the mean % inhibition ± the SEM. Wild-type N2 n = 9, mpz-1(tm1136) n = 10, mpz-1(gk273/+) n = 12 and mpz-1(tm1136/gk273) n = 12. There is no significant difference between the wild type and mutants (two-way ANOVA). Scale bar: x = 2.5 min, y = 3 mV
Discussion
Scaffolding proteins perform an important role in coordinating GPCR function (Romero et al. 2011). This can be in terms of subcellular targeting, localisation, immobilisation and specifying downstream signalling cascades through multi-meric protein networks. The metabotropic glutamate receptors are a class of GPCR whose function is scaffolded through interactions between the intracellular C-terminal and a variety of proteins (Enz 2012). Our current understanding of these scaffolding interactions suggests that they are important for modulating synaptic signalling and synaptic plasticity within neural networks, and their disruption can contribute to the mechanisms underlying CNS disorders such as Fragile-X (Ronesi and Huber 2008; Ronesi et al. 2012). In C. elegans, comparatively less is understood in terms of the identity and function of proteins that are involved in the scaffolding of GPCR function. This is surprising considering the worm has the largest number of genes (~ 1100) encoding GPCRs compared to humans and Drosophila (Bargmann 1998). Proteins that have emerged as scaffolding receptor function in C. elegans have been identified through either forwards genetic approaches (Byrd et al. 2001) or predictive approaches based upon known function in other organisms to screen candidates with the potential to participate in protein–protein interactions (Emtage et al. 2009; Danielson et al. 2012). In view of this limited understanding and the significant role that such mechanisms play, we used a yeast two-hybrid screen to identify protein–protein interactions that scaffold the C. elegans mGluR orthologue MGL-1.
The screen yielded three candidates that were prioritised for further analysis, MPZ-1, NRFL-1 and PTP-1. Each of the three contains one or more tandem PDZ-binding domains; this was particularly intriguing since the MGL receptor C-terminal contains a PDZ-binding motif. Based on the criteria of shared cellular expression, our observations would suggest that the NRFL-1 interaction is unlikely to have physiological relevance. Our reporters were designed to ape physiological expression, and in the case of NRFL-1, we seem to have a similar pattern to previously described (Hagiwara et al. 2012).
The mammalian orthologues of MPZ-1 and PTP-1 (MUPP-1 and PTPMEG, respectively) have each been identified as scaffolding receptor function. MUPP-1 was first identified as interacting with the 5-HT2C receptor. It has since been shown to interact with a range of different proteins, including ion channels and adhesion molecules. In general, it is considered that MUPP-1 serves to organise diverse signalling complexes (Jang et al. 2014; Fujita et al. 2012; Baumgart et al. 2014; Sindic et al. 2009), and it is has been shown that the disruption of MUPP-1 interactions can lead to the destabilisation of receptor coupling to G-proteins (Guillaume et al. 2008). In terms of the physiological function of MUPP-1, these are diverse, but perhaps the largest single body of work relates to its role in alcohol responses (Kruse et al. 2014; Milner et al. 2015; Metten et al. 2014; Buck et al. 2012; Karpyak et al. 2009) and the regulation of excitability during withdrawal. MPZ-1 has previously been shown to scaffold the function of distinct non-glutamatergic receptor types in C. elegans. MPZ-1 has been described as scaffolding the function of the 5-HT GPCR SER-1 (Xiao et al. 2006), and a molecular interaction has been identified with the insulin DAF-2 receptor (Palmitessa and Benovic 2010).
The other candidate, PTP-1, is particularly interesting because of the functional domains it contains, FERM, PDZ and PTP. PTP-1 has not been previously described as a scaffolding protein, and there is relatively little known about its function in C. elegans other than the FERM domain required for localisation in nerve processes (Uchida et al. 2002). Intriguingly, our study has identified that PTP-1 can interact with the C-terminal of the AMPA like receptor subunit GLR-1, which like MGL-1 contains a Type I PDZ motif. Indeed the mammalian orthologue of PTP-1, PTPMEG has been identified as binding to the intracellular C-terminal of the Gluδ2 ionotropic subunit, and it has been suggested that this interaction underlies mechanisms of synaptic plasticity in the cerebellum (Kohda et al. 2013). This modulation extends to the mammalian family of mGluRs, where protein phosphatase 1 (PP1) has been shown to interact with the intracellular C-termini of mGluR1a, mGluR5a/b and 7a (Croci et al. 2003; Meiselbach et al. 2006).
MPZ-1 and PTP-1 were both co-expressed with MGL-1 in neurones; in particular, we identified a substantial overlap between MPZ-1 and MGL-1 in the pharyngeal nervous system. Amongst other cells, co-expression in pharyngeal neurons is likely to include NSM and M4. NSM is serotonergic/glutamatergic and has been identified as performing a role in modulating dwelling and roaming behaviour, whereas M4 is cholinergic and controls isthmus peristalsis during feeding, respectively. Our analysis of strains carrying mutations in the mpz-1 gene identified no gross changes in pharyngeal responses to L-CCG-I, which provides a pharmacological readout for MGL-1 function. This may be a reflection of the limited contribution of the cells co-expressing MGL-1 and MPZ-1 to the network effect of L-CCG-I. We identified that MGL-1 is expressed in pharyngeal neurones independently of MPZ-1 and the function of MGL-1 in these cells may have a significant contribution to the L-CCG-I effect. Alternatively, a more discrete analysis is required to determine how MPZ-1 scaffolds the function of MGL-1 in the cells identified. Our transcript analysis of the mutants carrying the gk273 or tm1136 allele identified products corresponding to the mutant transcripts. MPZ-1 is a highly complex gene with a number of different 5′ isoforms, and 10 are currently annotated on Wormbase (Version WS253) (C52A11.4a-j). The possibility remains that protein translation could be initiated from the mutant transcripts at an alternative start codon downstream of the regions deleted. This would generate a 5′ truncated MPZ-1 protein with residual activity. Indeed, the isoform C52A11.4i is a 5′ truncated isoform that contains the 3′ region of MPZ-1 defined by the two-hybrid prey clone and could potentially interact with MGL-1.
The data from this study indicate a potent in vitro ability of the MGL-1 receptor to recruit scaffolding proteins. These can be multi-domain proteins that are involved in cross-linking and signal complex assembly, subcellular organisation (FERM domain) and impart discrete signalling (phosphatase activity). These findings hint at the integrative ability of the MGL receptors, evidenced from functional studies in higher organisms. The functional scaffolding of this class of receptor will play out in C. elegans, and the experimental tractability of this model organism will facilitate further investigation of such interactions for the modulation of neural networks and behaviour.
References
Azkue JJ, Mateos JM, Elezgarai I, Benítez R, Osorio A, Díez J et al (2000) The metabotropic glutamate receptor subtype mGluR 2/3 is located at extrasynaptic loci in rat spinal dorsal horn synapses. Neurosci Lett 287(3):236–238
Bargmann CI (1998) Neurobiology of the Caenorhabditis elegans genome. Science 282(5396):2028–2033
Baumgart S, Jansen F, Bintig W, Kalbe B, Herrmann C, Klumpers F et al (2014) The scaffold protein MUPP1 regulates odorant-mediated signaling in olfactory sensory neurons. J Cell Sci 127(Pt 11):2518–2527. https://doi.org/10.1242/jcs.144220
Blümcke I, Behle K, Malitschek B, Kuhn R, Knöpfel T, Wolf HK et al (1996) Immunohistochemical distribution of metabotropic glutamate receptor subtypes mGluR1b, mGluR2/3, mGluR4a and mGluR5 in human hippocampus. Brain Res 736(1–2):217–226
Breeden L, Nasmyth K (1985) Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol 50:643–650
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77(1):71–94
Buck KJ, Milner LC, Denmark DL, Grant SG, Kozell LB (2012) Discovering genes involved in alcohol dependence and other alcohol responses: role of animal models. Alcohol Res 34(3):367–374
Byrd DT, Kawasaki M, Walcoff M, Hisamoto N, Matsumoto K, Jin Y (2001) UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron 32(5):787–800
Conn PJ, Pin JP (1997) Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol 37:205–237. https://doi.org/10.1146/annurev.pharmtox.37.1.205
Croci C, Sticht H, Brandstätter JH, Enz R (2003) Group I metabotropic glutamate receptors bind to protein phosphatase 1C. Mapping and modeling of interacting sequences. J Biol Chem 278(50):50682–50690. https://doi.org/10.1074/jbc.m305764200
Danielson E, Zhang N, Metallo J, Kaleka K, Shin SM, Gerges N et al (2012) S-SCAM/MAGI-2 is an essential synaptic scaffolding molecule for the GluA2-containing maintenance pool of AMPA receptors. J Neurosci 32(20):6967–6980. https://doi.org/10.1523/JNEUROSCI.0025-12.2012
Dillon J, Franks CJ, Murray C, Edwards RJ, Calahorro F, Ishihara T et al (2015) Metabotropic glutamate receptors: modulators of context-dependent feeding behaviour in C. elegans. J Biol Chem 290(24):15052–15065. https://doi.org/10.1074/jbc.M114.606608
Emtage L, Chang H, Tiver R, Rongo C (2009) MAGI-1 modulates AMPA receptor synaptic localization and behavioral plasticity in response to prior experience. PLoS ONE 4(2):e4613. https://doi.org/10.1371/journal.pone.0004613
Enz R (2012) Structure of metabotropic glutamate receptor C-terminal domains in contact with interacting proteins. Front Mol Neurosci 5:52. https://doi.org/10.3389/fnmol.2012.00052
Fujita E, Tanabe Y, Imhof BA, Momoi MY, Momoi T (2012) A complex of synaptic adhesion molecule CADM1, a molecule related to autism spectrum disorder, with MUPP1 in the cerebellum. J Neurochem 123(5):886–894. https://doi.org/10.1111/jnc.12022
Guillaume JL, Daulat AM, Maurice P, Levoye A, Migaud M, Brydon L et al (2008) The PDZ protein mupp1 promotes Gi coupling and signaling of the Mt1 melatonin receptor. J Biol Chem 283(24):16762–16771. https://doi.org/10.1074/jbc.M802069200
Hagiwara K, Nagamori S, Umemura YM, Ohgaki R, Tanaka H, Murata D, Nakagomi S, Nomura KH, Kage-Nakadai E, Mitani S, Nomura K, Kanai Y (2012) NRFL-1, the C. elegans NHERF orthologue, interacts with amino acid transporter 6 (AAT-6) for age-dependent maintenance of AAT-6 on the membrane. PLoS ONE 7(8):e43050. https://doi.org/10.1371/journal.pone.0043050
Jang WH, Choi SH, Jeong JY, Park JH, Kim SJ, Seog DH (2014) Neuronal cell-surface protein neurexin 1 interaction with multi-PDZ domain protein MUPP1. Biosci Biotechnol Biochem 78(4):644–646. https://doi.org/10.1080/09168451.2014.890031
Jeong H, Paik YK (2017) MGL-1 on AIY neurons translates starvation to reproductive plasticity via neuropeptide signaling in Caenorhabditis elegans. Dev Biol 430(1):80–89. https://doi.org/10.1016/j.ydbio.2017.08.014
Karpyak VM, Kim JH, Biernacka JM, Wieben ED, Mrazek DA, Black JL et al (2009) Sequence variations of the human MPDZ gene and association with alcoholism in subjects with European ancestry. Alcohol Clin Exp Res 33(4):712–721. https://doi.org/10.1111/j.1530-0277.2008.00888.x
Kohda K, Kakegawa W, Yuzaki M (2013) Unlocking the secrets of the δ2 glutamate receptor: a gatekeeper for synaptic plasticity in the cerebellum. Commun Integr Biol 6(6):e26466. https://doi.org/10.4161/cib.26466
Kruse LC, Walter NA, Buck KJ (2014) Mpdz expression in the caudolateral substantia nigra pars reticulata is crucially involved in alcohol withdrawal. Genes Brain Behav 13(8):769–776. https://doi.org/10.1111/gbb.12171
Lujan R, Nusser Z, Roberts JD, Shigemoto R, Somogyi P (1996) Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur J Neurosci 8(7):1488–1500
Luján R, Roberts JD, Shigemoto R, Ohishi H, Somogyi P (1997) Differential plasma membrane distribution of metabotropic glutamate receptors mGluR1 alpha, mGluR2 and mGluR5, relative to neurotransmitter release sites. J Chem Neuroanat 13(4):219–241
Meiselbach H, Sticht H, Enz R (2006) Structural analysis of the protein phosphatase 1 docking motif: molecular description of binding specificities identifies interacting proteins. Chem Biol 13(1):49–59. https://doi.org/10.1016/j.chembiol.2005.10.009
Metten P, Iancu OD, Spence SE, Walter NA, Oberbeck D, Harrington CA et al (2014) Dual-trait selection for ethanol consumption and withdrawal: genetic and transcriptional network effects. Alcohol Clin Exp Res 38(12):2915–2924. https://doi.org/10.1111/acer.12574
Milner LC, Shirley RL, Kozell LB, Walter NA, Kruse LC, Komiyama NH et al (2015) Novel MPDZ/MUPP1 transgenic and knockdown models confirm Mpdz’s role in ethanol withdrawal and support its role in voluntary ethanol consumption. Addict Biol 20(1):143–147. https://doi.org/10.1111/adb.12087
Nusser Z, Mulvihill E, Streit P, Somogyi P (1994) Subsynaptic segregation of metabotropic and ionotropic glutamate receptors as revealed by immunogold localization. Neuroscience 61(3):421–427
Palmitessa A, Benovic JL (2010) Arrestin and the multi-PDZ domain-containing protein MPZ-1 interact with phosphatase and tensin homolog (PTEN) and regulate Caenorhabditis elegans longevity. J Biol Chem 285(20):15187–15200. https://doi.org/10.1074/jbc.M110.104612
Perroy J, Gutierrez GJ, Coulon V, Bockaert J, Pin JP, Fagni L (2001) The C terminus of the metabotropic glutamate receptor subtypes 2 and 7 specifies the receptor signaling pathways. J Biol Chem 276(49):45800–45805. https://doi.org/10.1074/jbc.M106876200
Riedel G, Reymann KG (1996) Metabotropic glutamate receptors in hippocampal long-term potentiation and learning and memory. Acta Physiol Scand 157(1):1–19. https://doi.org/10.1046/j.1365-201X.1996.484231000.x
Riedel G, Wetzel W, Reymann KG (1996) Comparing the role of metabotropic glutamate receptors in long-term potentiation and in learning and memory. Prog Neuropsychopharmacol Biol Psychiatry 20(5):761–789
Romero G, von Zastrow M, Friedman PA (2011) Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: means, motif, and opportunity. Adv Pharmacol 62:279–314. https://doi.org/10.1016/B978-0-12-385952-5.00003-8
Ronesi JA, Huber KM (2008) Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci 28(2):543–547. https://doi.org/10.1523/JNEUROSCI.5019-07.2008
Ronesi JA, Collins KA, Hays SA, Tsai NP, Guo W, Birnbaum SG et al (2012) Disrupted Homer scaffolds mediate abnormal mGluR5 function in a mouse model of fragile X syndrome. Nat Neurosci 15(3):431–440. https://doi.org/10.1038/nn.3033
Shigemoto R, Kulik A, Roberts JD, Ohishi H, Nusser Z, Kaneko T et al (1996) Target-cell-specific concentration of a metabotropic glutamate receptor in the presynaptic active zone. Nature 381(6582):523–525. https://doi.org/10.1038/381523a0
Sindic A, Huang C, Chen AP, Ding Y, Miller-Little WA, Che D et al (2009) MUPP1 complexes renal K + channels to alter cell surface expression and whole cell currents. Am J Physiol Renal Physiol 297(1):F36–F45. https://doi.org/10.1152/ajprenal.90559.2008
Uchida Y, Ogata M, Mori Y, Oh-hora M, Hatano N, Hamaoka T (2002) Localization of PTP-FERM in nerve processes through its FERM domain. Biochem Biophys Res Commun 292(1):13–19
Xiao H, Hapiak VM, Smith KA, Lin L, Hobson RJ, Plenefisch J et al (2006) SER-1, a Caenorhabditis elegans 5-HT2-like receptor, and a multi-PDZ domain containing protein (MPZ-1) interact in vulval muscle to facilitate serotonin-stimulated egg-laying. Dev Biol 298(2):379–391. https://doi.org/10.1016/j.ydbio.2006.06.044
Acknowledgements
Strains were provided by either the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) or provided by the MITANI Laboratory through the National Bio-Resource Project of the MEXT, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Dillon, J., Holden-Dye, L. & O’Connor, V. Yeast two-hybrid screening identifies MPZ-1 and PTP-1 as candidate scaffolding proteins of metabotropic glutamate receptors in Caenorhabditis elegans. Invert Neurosci 18, 16 (2018). https://doi.org/10.1007/s10158-018-0218-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10158-018-0218-2