Abstract
Background
BRAF V600E is a common mutation in melanoma, and BRAF inhibitors are effective in treating of BRAF mutation-positive melanoma. DNA carrying this mutation is released from melanoma cells into the circulation. As such, circulating tumor-derived DNA (ctDNA) in peripheral blood represents a novel biomarker for evaluating tumor features in cancer patients. However, ctDNA is present in the peripheral blood at very low levels, which makes the detection of specific mutations in this DNA a challenge. Competitive allele-specific TaqMan PCR (castPCR), a straightforward commercially available assay, is a sensitive technique for quantitating a small amount of DNA.
Methods
The level of BRAF V600E ctDNA was quantified by castPCR in 26 consecutive plasma samples from six melanoma patients.
Results
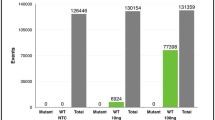
The castPCR assay was performed using a mixture of BRAF V600E DNA and BRAF wild DNA and found to be able to detect BRAF V600E at a fractional abundance of ≥0.5 % in 2- to 10-ng samples of genomic DNA. Cell-free DNA was then extracted from peripheral blood samples collected from six patients with melanoma harboring the BRAF V600E mutation. BRAF V600E ctDNA was detected in three patients, at a fractional abundance of between 1.28 and 58.0 % of total BRAF cell-free DNA. The abundance of BRAF V600E ctDNA correlated with tumor burden, as determined by computed tomography imaging. In two cases, an increase in the level of BRAF V600E ctDNA preceded exacerbation of clinical symptoms.
Conclusion
The castPCR assay can detect and quantitate small amounts of BRAF V600E ctDNA in samples containing large amounts of BRAF wild cell-free DNA. Thus, we suggest that the castPCR assay is suitable for monitoring ctDNA in the plasma of melanoma patients.




Similar content being viewed by others
References
Menzies AM, Long GV (2014) Systemic treatment for BRAF-mutant melanoma: where do we go next? Lancet Oncol 15:e371–e381
Diaz-Lagares A, Alegre E, Arroyo A et al (2011) Evaluation of multiple serum markers in advanced melanoma. Tumour Biol 32:1155–1161
Sanmamed MF, Fernandez-Landazuri S, Rodriguez C et al (2014) Relevance of MIA and S100 serum tumor markers to monitor BRAF inhibitor therapy in metastatic melanoma patients. Clin Chim Acta 429:168–174
Schwarzenbach H, Hoon DS, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11:426–437
Dawson SJ, Tsui DW, Murtaza M et al (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368:1199–1209
Oxnard GR, Paweletz CP, Kuang Y et al (2014) Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 20:1698–1705
Taly V, Pekin D, Benhaim L et al (2013) Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem 59:1722–1731
Thierry AR, Mouliere F, El Messaoudi S et al (2014) Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 20:430–435
Huggett JF, Whale A (2013) Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin Chem 59:1691–1693
Siravegna G, Bardelli A (2014) Minimal residual disease in breast cancer: in blood veritas. Clin Cancer Res 20:2505–2507
Ashida A, Uhara H, Kiniwa Y et al (2012) Assessment of BRAF and KIT mutations in Japanese melanoma patients. J Dermatol Sci 66:240–242
Cheng SP, Hsu YC, Liu CL et al (2014) Significance of allelic percentage of BRAF c.1799T > A (V600E) mutation in papillary thyroid carcinoma. Ann Surg Oncol 21[Suppl 4]:S619–S626
Didelot A, Le Corre D, Luscan A et al (2012) Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR mutation detection in clinical formalin fixed paraffin embedded samples. Exp Mol Pathol 92:275–280
Ashida A, Uhara H, Mikoshiba A et al (2015) Melanoma with BRAF mutation in circulating cell-free DNA despite no mutation in the primary lesion: a case report. Acta Derm Venereol 96:128–129
Schreuer M, Meersseman G, van Den Herrewegen S et al (2016) Applications for quantitative measurement of BRAF V600 mutant cell-free tumor DNA in the plasma of patients with metastatic melanoma. Melanoma Res 26:157–163
Wakamatsu K, Ito S, Horikoshi T (1991) Normal values of urinary excretion and serum concentration of 5-S-cysteinyldopa and 6-hydroxy-5-methoxyindole-2-carboxylic acid, biochemical markers of melanoma progression. Melanoma Res 1:141–147
Sakaizawa K, Goto Y, Kiniwa Y et al (2012) Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br J Cancer 106:939–946
Stoecklein NH, Hosch SB, Bezler M et al (2008) Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell 13:441–453
Diehl F, Schmidt K, Choti MA et al (2008) Circulating mutant DNA to assess tumor dynamics. Nat Med 14:985–990
Taniguchi K, Uchida J, Nishino K et al (2011) Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 17:7808–7815
Sanmamed MF, Fernandez-Landazuri S, Rodriguez C et al (2015) Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clin Chem 61:297–304
Kukita Y, Uchida J, Oba S et al (2013) Quantitative identification of mutant alleles derived from lung cancer in plasma cell-free DNA via anomaly detection using deep sequencing data. PLoS One 8:e81468
Wakamatsu K, Kageshita T, Furue M et al (2002) Evaluation of 5-S-cysteinyldopa as a marker of melanoma progression: 10 years’ experience. Melanoma Res 12:245–253
Colombino M, Capone M, Lissia A et al (2012) BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol 30:2522–2529
Saint-Jean M, Quereux G, Nguyen JM et al (2014) Is a single BRAF wild-type test sufficient to exclude melanoma patients from vemurafenib therapy? J Invest Dermatol 134:1468–1470
Acknowledgments
We thank Dr. Wakamatsu of the Fujita Health University School of Health Sciences for measurements of 5-SCD. This study was supported by a Grant-in-Aid for Scientific Research (C) 26461687 (to A. A.) from Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
About this article
Cite this article
Ashida, A., Sakaizawa, K., Mikoshiba, A. et al. Quantitative analysis of the BRAF V600E mutation in circulating tumor-derived DNA in melanoma patients using competitive allele-specific TaqMan PCR. Int J Clin Oncol 21, 981–988 (2016). https://doi.org/10.1007/s10147-016-0976-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-016-0976-y