Abstract
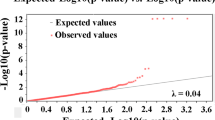
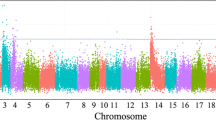
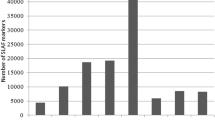
Selection of new lines with high salinity tolerance allows for economically feasible production of tilapias in brackish water areas. Mapping QTLs and identifying the markers linked to salinity-tolerant traits are the first steps in the improvement of the tolerance in tilapia through marker-assisted selection techniques. By using QTL-seq strategy and linkage-based analysis, two significant QTL intervals (chrLG4 and chrLG18) on salinity-tolerant traits were firstly identified in the Nile tilapia. Fine mapping with microsatellite and SNP markers suggested a major QTL region that located at 23.0 Mb of chrLG18 and explained 79% of phenotypic variation with a LOD value of 95. Expression analysis indicated that at least 10 genes (e.g., LACTB2, KINH, NCOA2, DIP2C, LARP4B, PEX5R, and KCNJ9) near or within the QTL interval were significantly differentially expressed in intestines, brains, or gills under 10, 15, or 20 ppt challenges. Our findings suggest that QTL-seq can be effectively utilized in QTL mapping of salinity-tolerant traits in fish. The identified major QTL is a promising locus to improve our knowledge on the genetic mechanism of salinity tolerance in tilapia.




Similar content being viewed by others
References
Aruna A, Nagarajan G, Chang CF (2012) Involvement of corticotrophin-releasing hormone and corticosteroid receptors in the brain-pituitary-gill of tilapia during the course of seawater acclimation. J Neuroendocrinol 24(5):818–830
Avarre JC, Dugue R, Alonso P, Diombokho A, Joffrois C, Faivre N, Cochet C, Durand JD (2014) Analysis of the black-chinned tilapia Sarotherodon melanotheron heudelotii reproducing under a wide range of salinities: from RNA-seq to candidate genes. Mol Ecol Resour 14(1):139–149
Bras YL, Dechamp N, Krieg F, Filangi O, Guyomard R, Boussaha M, Bovenhuis H, Pottinger TG, Prunet P, Roy PL (2011) Detection of QTL with effects on osmoregulation capacities in the rainbow trout (Oncorhynchus mykiss). BMC Genet 12(1):46
Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, Turner-Maier J, Johnson J, Alcazar R, Noh HJ, Russell P, Aken B, Alfoldi J, Amemiya C, Azzouzi N, Baroiller JF, Barloy-Hubler F, Berlin A, Bloomquist R, Carleton KL, Conte MA, D’Cotta H, Eshel O, Gaffney L, Galibert F, Gante HF, Gnerre S, Greuter L, Guyon R, Haddad NS, Haerty W, Harris RM, Hofmann HA, Hourlier T, Hulata G, Jaffe DB, Lara M, Lee AP, MacCallum I, Mwaiko S, Nikaido M, Nishihara H, Ozouf-Costaz C, Penman DJ, Przybylski D, Rakotomanga M, Renn SCP, Ribeiro FJ, Ron M, Salzburger W, Sanchez-Pulido L, Santos ME, Searle S, Sharpe T, Swofford R, Tan FJ, Williams L, Young S, Yin S, Okada N, Kocher TD, Miska EA, Lander ES, Venkatesh B, Fernald RD, Meyer A, Ponting CP, Streelman JT, Lindblad-Toh K, Seehausen O, Di Palma F (2014) The genomic substrate for adaptive radiation in African cichlid fish. Nature 513(7518):375–381
Bystriansky JS, Richards JG, Schulte PM, Ballantyne JS (2006) Reciprocal expression of gill Na+/K+-ATPase alpha-subunit isoforms alpha1a and alpha1b during seawater acclimation of three salmonid fishes that vary in their salinity tolerance. J Exp Biol 209(10):1848–1858
Cnaani A, Zilberman N, Tinman S, Hulata G, Ron M (2004) Genome-scan analysis for quantitative trait loci in an F-2 tilapia hybrid. Mol Gen Genomics 272:162–172
Das S, Upadhyaya HD, Bajaj D, Kujur A, Badoni S, Laxmi, Kumar V, Tripathi S, Gowda CL, Sharma S, Singh S, Tyagi AK, Parida SK (2015) Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea. DNA Res 22(3):193–203
Dean DB, Whitlow ZW, Borski RJ (2003) Glucocorticoid receptor upregulation during seawater adaptation in a euryhaline teleost, the tilapia (Oreochromis mossambicus). Gen Comp Endocrinol 132(1):112–118
Eknath AE, Hulata G (2009) Use and exchange of genetic resources of Nile tilapia (Oreochromisƒniloticus). Rev Aquac 1(3-4):197–213
Evans TG (2010) Co-ordination of osmotic stress responses through osmosensing and signal transduction events in fishes. J Fish Biol 76(8):1903–1925
Evans D, Claiborne J (2006) The physiology of fishe, 3rd edn. CRC, Boca Raton
Evans DH, Piermarini PM, Choe KP (2004) The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85:97
Farcy E, Serpentini A, Fievet B, Lebel JM (2007) Identification of cDNAs encoding HSP70 and HSP90 in the abalone Haliotis tuberculata: transcriptional induction in response to thermal stress in hemocyte primary culture. Comp Biochem Physiol B Biochem Mol Biol 146(4):540–550
Fiol DF, Sanmarti E, Lim AH, Kültz D (2011) A novel GRAIL E3 ubiquitin ligase promotes environmental salinity tolerance in euryhaline tilapia. Biochim Biophys Acta Gen Subj 1810(4):439–445
Gil J, Peters G (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7(9):667–677
Gu XH, Li BJ, Lin HR, Xia JH (2018) Unraveling the associations of the tilapia DNA polymerase delta subunit 3 (POLD3) gene with saline tolerance traits. Aquaculture 485:53–58
Haupt Y, Bath ML, Harris AW, Adams JM (1993) bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene 8(11):3161–3164
Hiroi J, Mccormick SD, Ohtani-Kaneko R, Kaneko T (2005) Functional classification of mitochondrion-rich cells in euryhaline Mozambique tilapia (Oreochromis mossambicus) embryos, by means of triple immunofluorescence staining for Na+/K+-ATPase, Na+/K+/2Cl− cotransporter and CFTR anion channel. J Exp Biol 208(11):2023–2036
Hiroi J, Yasumasu S, Mccormick SD, Hwang PP, Kaneko T (2008) Evidence for an apical Na-Cl cotransporter involved in ion uptake in a teleost fish. J Exp Biol 211(16):2584–2599
Howe AE (2004) The genetic basis of red color in tilapia. M.S. thesis, Univ. of New Hampshire, Durham
Huang CW, Li YH, Hu SY, Chi JR, Lin GH, Lin CC, Gong HY, Chen JY, Chen RH, Chang SJ, Liu FG, Wu JL (2012) Differential expression patterns of growth-related microRNAs in the skeletal muscle of Nile tilapia (Oreochromis niloticus). J Anim Sci 90(12):4266–4279
Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M (1999) Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13(20):2678–2690
Kamal AHMM, Mair GC (2005) Salinity tolerance in superior genotypes of tilapia, Oreochromis niloticus, Oreochromis mossambicus and their hybrids. Aquaculture 247(1-4):189–201
Katagiri T, Kidd C, Tomasino E, Davis JT, Wishon C, Stern JE, Carleton KL, Howe AE, Kocher TD (2005) A BAC-based physical map of the Nile tilapia genome. BMC Genomics 6(1):89
Kiilerich P, Kristiansen K, Madsen SS (2007) Cortisol regulation of ion transporter mRNA in Atlantic salmon gill and the effect of salinity on the signaling pathway. J Endocrinol 194(2):417–427
Kofler R, Pandey RV, Schlotterer C (2011) PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27(24):3435–3436
Kültz D (2012) The combinatorial nature of osmosensing in fishes. Physiology 27(4):259–275
Kultz D, Li J, Gardell A, Sacchi R (2013) Quantitative molecular phenotyping of gill remodeling in a Cichlid fish responding to salinity stress. Mol Cell Proteomics 12(12):3962–3975
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359
Lee K, Adhikary G, Balasubramanian S, Gopalakrishnan R, McCormick T, Dimri GP, Eckert RL, Rorke EA (2008) Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol 128(1):9–17
Li BJ, Li HL, Meng Z, Zhang Y, Lin H, Yue GH, Xia JH (2017a) Copy number variations in tilapia genomes. Mar Biotechnol (NY). 19(1):11–21
Li HL, Gu XH, Li BJ, Chen CH, Lin HR, Xia JH (2017b) Genome-wide QTL analysis identified significant associations between hypoxia tolerance and mutations in the GPR132 and ABCG4 genes in Nile tilapia. Mar Biotechnol (NY) 19(5):441–453
Liu ZJ, Cordes JF (2004) DNA marker technologies and their applications in aquaculture genetics. Aquaculture 242(1-4):735–736
Liu F, Sun F, Li J, Xia JH, Lin G, Tu RJ, Yue GH (2013) A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus x Oreochromis spp.) and mapping of sex-determining loci. BMC Genomics 14(1):58
Lorin-Nebel C, Avarre JC, Faivre N, Wallon S, Charmantier G, Durand JD (2012) Osmoregulatory strategies in natural populations of the black-chinned tilapia Sarotherodon melanotheron exposed to extreme salinities in West African estuaries. J Comp Physiol B 182(6):771–780
McConnell SK, Beynon C, Leamon J, Skibinski DO (2000) Microsatellite marker based genetic linkage maps of Oreochromis aureus and O. niloticus (Cichlidae): extensive linkage group segment homologies revealed. Anim Genet 31(3):214–218
McCormick SD (2001) Endocrine control of osmoregulation in teleost fish. Am Zool 41:781–794
Mccormick SD, Regish A, O’Dea MF, Shrimpton JM (2008) Are we missing a mineralocorticoid in teleost fish? Effects of cortisol, deoxycorticosterone and aldosterone on osmoregulation, gill Na+,K+ -ATPase activity and isoform mRNA levels in Atlantic salmon. Gen Comp Endocrinol 157(1):35–40
Mccormick SD, Regish AM, Christensen AK (2009) Distinct freshwater and seawater isoforms of Na+/K+-ATPase in gill chloride cells of Atlantic salmon. J Exp Biol 212(24):3994–4001
Moen T, Agresti JJ, Cnaani A, Moses H, Famula TR, Hulata G, Gall GAE, May B (2004) A genome scan of a four-way tilapia cross supports the existence of a quantitative trait locus for cold tolerance on linkage group 23. Aquac Res 35(9):893–904
Norman JD, Danzmann RG, Glebe B, Ferguson MM (2011) The genetic basis of salinity tolerance traits in Arctic charr (Salvelinus alpinus). BMC Genet 12(1):81
Norman JD, Robinson M, Glebe B, Ferguson MM, Danzmann RG (2012) Genomic arrangement of salinity tolerance QTL in salmonids: a comparative analysis of Atlantic salmon (Salmo salar) with Arctic charr (Salvelinus alpinus) and rainbow trout (Oncorhynchus mykiss). BMC Genomics 13(1):420
Norman JD, Ferguson MM, Danzmann RG (2014) Transcriptomics of salinity tolerance capacity in Arctic charr (Salvelinus alpinus): a comparison of gene expression profiles between divergent QTL genotypes. Physiol Genomics 46(4):123–137
Palaiokostas C, Bekaert M, Khan MG, Taggart JB, Gharbi K, McAndrew BJ, Penman DJ (2013) Mapping and validation of the major sex-determining region in Nile tilapia (Oreochromis niloticus L.) using RAD sequencing. PLoS One 8(7):e68389
Rengmark, A.H. and F. Lingaas (2007) Genomic structure of the Nile tilapia (Oreochromis niloticus) transferrin gene and a haplotype associated with saltwater tolerance. Aquaculture, 272:146–155
Richards JG, Semple JW, Bystriansky JS, Schulte PM (2003) Na+/K+-ATPase alpha-isoform switching in gills of rainbow trout (Oncorhynchus mykiss) during salinity transfer. J Exp Biol 206(24):4475–4486
Sakamoto T, McCormick SD (2006) Prolactin and growth hormone in fish osmoregulation. Gen Comp Endocrinol 147(1):24–30
Seale AP, Fiess JC, Hirano T, Cooke IM, Grau EG (2006) Disparate release of prolactin and growth hormone from the tilapia pituitary in response to osmotic stimulation. Gen Comp Endocrinol 145(3):222–231
Smith HW (1930) The absorption and excretion of water and salts by marine teleosts. Am J Physiol 93:480
Stauffer JR Jr, Van DK, Hocutt CH (1984) Effects of salinity on preferred and lethal temperatures of the blackchin tilapia, Sarotherodon melanotheron. Water Resour Bull 20(5):771–775
Streelman JT, Kocher TD (2002) Microsatellite variation associated with prolactin expression and growth of salt-challenged tilapia. Physiol Genomics 9(1):1–4
Sun YB (2017) FasParser: a package for manipulating sequence data. Zool Res 38(2):110–112
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74(1):174–183
Thomsen DS, Anders K, Nielsen EE, Larsen PF, Olsvik PA, Volker L (2008) Interpopulation differences in expression of candidate genes for salinity tolerance in winter migrating anadromous brown trout (Salmo trutta L.) BMC Genet 9:12
Tipsmark CK, Madsen SS, Seidelin M, Christensen AS, Cutler CP, Cramb G (2002) Dynamics of Na(+),K(+),2Cl(−) cotransporter and Na(+),K(+)-ATPase expression in the branchial epithelium of brown trout (Salmo trutta) and Atlantic salmon (Salmo salar). J Exp Zool 293(2):106–118
Tipsmark CK, Kiilerich P, Nilsen TO, Ebbesson LO, Stefansson SO, Madsen SS (2008) Branchial expression patterns of claudin isoforms in Atlantic salmon during seawater acclimation and smoltification. Am J Physiol Regul Integr Comp Physiol 294(5):R1563–R1574
Van Bers NE, Crooijmans RP, Groenen MA, Dibbits BW, Komen J (2012) SNP marker detection and genotyping in tilapia. Mol Ecol Resour 12(5):932–941
Van Ooijen JW, 2009. MapQTL 6.0, software for the mapping of quantitative trait loci in experimental populations of dihaploid species
Van Ooijen JW (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93(05):343–349
Wang X, Kultz D (2017) Osmolality/salinity-responsive enhancers (OSREs) control induction of osmoprotective genes in euryhaline fish. Proc Natl Acad Sci U S A 114(13):E2729–E2738
Watanabe W, Kuo C-M, Huang M-C (1985a) Salinity tolerance of Nile tilapia (Oreochromis niloticus) spawned and hatched at various salinities. Aquaculture 48(2):159–176
Watanabe WO, Kuo C-M, Huang M-C (1985b) The ontogeny of salinity tolerance in the tilapias Oreochromis aureus, O. niloticus, and an O. mossambicus × O. niloticus hybrid, spawned and reared in freshwater. Aquaculture 47(4):353–367
Xia JH, Wan ZY, Ng ZL, Wang L, Fu GH, Lin G, Liu F, Yue GH (2014) Genome-wide discovery and in silico mapping of gene-associated SNPs in Nile tilapia. Aquaculture 432:67–73
Xia JH, Bai Z, Meng Z, Zhang Y, Wang L, Liu F, Jing W, Wan ZY, Li J, Lin H, Yue GH (2015) Signatures of selection in tilapia revealed by whole genome resequencing. Sci Rep 5:14168
Xu Z, Gan L, Li T, Xu C, Chen K, Wang X, Qin JG, Chen L, Li E (2015) Transcriptome profiling and molecular pathway analysis of genes in association with salinity adaptation in Nile tilapia Oreochromis niloticus. PLoS One 10(8):e0136506
Yan B, Guo JT, Zhao LH, Zhao JL (2012) MiR-30c: a novel regulator of salt tolerance in tilapia. Biochem Biophys Res Commun 425(2):315–320
Yue GH (2014) Recent advances of genome mapping and marker-assisted selection in aquaculture. Fish Fish 15(3):376–396
Yue GH, Lin H, Li J (2016) Tilapia is the fish for next-generation aquaculture. Int J Marine Sci Ocean Technol 3:11–13
Zhang R, Zhang LL, Ye X, Tian YY, Sun CF, Lu MX, Bai JJ (2013) Transcriptome profiling and digital gene expression analysis of Nile tilapia (Oreochromis niloticus) infected by Streptococcus agalactiae. Mol Biol Rep 40(10):5657–5668
Zhang M, Sun Y, Liu Y, Qiao F, Chen L, Liu W-T, Du Z, Li E (2016) Response of gut microbiota to salinity change in two euryhaline aquatic animals with reverse salinity preference. Aquaculture 454:72–80
Acknowledgements
We thank two anonymous reviewers for comments on the manuscript. This work was supported by the National Natural Science Foundation of China (No. 31572612); Science and Technology Planning Project of Guangdong Province, China (2017A030303008); and Modern Agriculture Talents Support Program (2016–2020).
Author information
Authors and Affiliations
Contributions
JHX and HRL contributed to project conception. Experiment and data analysis was conducted by XHG, DLJ, HY, CCH, and BJL. The manuscript was prepared by JHX and DLJ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Electronic Supplementary Material
Supplementary Table S1
The primer list used for genotyping and qRT-PCR analysis. (XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Gu, X.H., Jiang, D.L., Huang, Y. et al. Identifying a Major QTL Associated with Salinity Tolerance in Nile Tilapia Using QTL-Seq. Mar Biotechnol 20, 98–107 (2018). https://doi.org/10.1007/s10126-017-9790-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-017-9790-4