Abstract
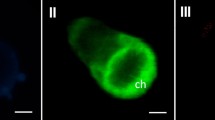
Transcription factors play a central role in expression of genomic information in all organisms. The objective of our study is to analyze the function of transcription factors in red algae. One way to analyze transcription factors in eukaryotic cells is to study their nuclear localization, as reported for land plants and green algae using fluorescent proteins. There is, however, no report documenting subcellular localization of transcription factors from red algae. In the present study, using the marine red alga Porphyra yezoensis, we confirmed for the first time successful expression of humanized fluorescent proteins (ZsGFP and ZsYFP) from a reef coral Zoanthus sp. and land plant-adapted sGFP(S65T) in gametophytic cells comparable to expression of AmCFP. Following molecular cloning and characterization of transcription factors DP-E2F-like 1 (PyDEL1), transcription elongation factor 1 (PyElf1) and multiprotein bridging factor 1 (PyMBF1), we then demonstrated that ZsGFP and AmCFP can be used to visualize nuclear localization of PyElf1 and PyMBF1. This is the first report to perform visualization of subcellular localization of transcription factors as genome-encoded proteins in red algae.






Similar content being viewed by others
References
Alexandre CM, Hennig L (2008) FLC or not FLC: the other side of vernalization. J Exp Bot 59:1127–1135
Asamizu E, Nakajima M, Kitade Y, Saga N, Nakamura Y, Tabata S (2003) Comparison of RNA expression profiles between the two generations of Porphyra yezoensis (Rhodophyta), based on expressed sequence tag frequency analysis. J Phycol 39:923–930
Babinger K, Hallmann A, Schmitt R (2006) Translational control of regA, a key gene controlling cell differentiation in Volvox carteri. Development 133:4045–4051
Bisova K, Krylov DM, Umen JG (2005) Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol 137:475–479
Brendel C, Gelman L, Auwerx J (2002) Multiprotein bridging factor-1 (MBF-1) is a cofactor for nuclear receptors that regulate lipid metabolism. Mol Endocrinol 16:1367–1377
Camuzeaux B, Spriet C, Héliot L, Coll J, Duterque-Coquillaud M (2005) Imaging Erg and Jun transcription factor interaction in living cells using fluorescence resonance energy transfer analyses. Biochem Biophys Res Commun 332:1107–1114
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451
Di Stefano L, Jensen MR, Helin K (2003) E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J 22:6289–6298
Fukuda S, Mikami K, Uji T, Park EJ, Ohba T, Asada K, Kitade Y, Endo H, Kato I, Saga N (2008) Factors influencing efficiency of transient gene expression in the red macrophyte Porphyra yezoensis. Plant Sci 174:329–339
Goodenough U, Lin H, Lee JH (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol 18:350–361
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Kosugi S, Ohashi Y (2002) E2Ls, E2F-like repressors of Arabidopsis that bind to E2F. J Biol Chem 277:16553–16558
Li J, Li X, Guo L, Lu F, Feng X, He K, Wei L, Chen Z, Qu LJ, Gu H (2006) A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J Exp Bot 57:1263–1273
Li GL, Roy B, Li XH, Yue WF, Wu XF, Liu JM, Zhang CX, Miao YG (2008) Quantification of silkworm coactivator of MBF1 mRNA by SYBR Green I real-time RT-PCR reveals tissue- and stage-specific transcription levels. Mol Biol Rep 36:1217–1223
Liu QX, Jindra M, Ueda H, Hiromi Y, Hirose S (2003) Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development 130:719–728
Liu QX, Nakashima-Kamimura N, Ikeo K, Hirose S, Gojobori T (2007) Compensatory change of interacting amino acids in the coevolution of transcriptional coactivator MBF1 and TATA-box-binding protein. Mol Biol Evol 24:1458–1463
Malone E, Fassler JS, Winston F (1993) Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet 237:449–459
Mikami K, Uji T, Li L, Takahashi M, Yasui H, Saga N (2009) Visualization of Phosphoinositides via the development of the transient expression system of a cyan fluorescent protein in the red alga Porphyra yezoensis. Mar Biotechnol doi:10.1007/s10126-008-9172-z
Nikaido I, Asamizu E, Nakajima M, Nakamura Y, Saga N, Tabata S (2000) Generation of 10, 154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res 7:223–227
Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18:455–463
Nowak K, Luniak N, Meyer S, Schulze J, Mendel RR, Hänsch R (2004) Fluorescent proteins in poplar: a useful tool to study promoter function and protein localization. Plant Biol 6:65–73
Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R (2002) Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J 31:319–330
Prather D, Krogan NJ, Emili A, Greenblatt JF, Winston F (2005) Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol Cell Biol 25:10122–35
Qian X, Gozani SN, Yoon H, Jeon CJ, Agarwal K, Weiss MA (1993) Novel zinc finger motif in the basal transcriptional machinery: three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS. Biochemistry 32:9944–9959
Reines D, Conaway JW, Conaway RC (1996) The RNA polymerase II general elongation factors. Trends Biochem Sci 21:351–355
Robbens S, Khadaroo B, Camasses A, Derelle E, Ferraz C, Inzé D, Van de Peer Y, Moreau H (2005) Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Mol Biol Evol 22:589–597
Saga N, Kitade Y (2002) Porphyra: a model plant in marine sciences. Fish Sci 68(Suppl):1075–1078
Saleh A, Lumbreras V, Lopez C, Dominguez-Puigjaner E, Kizis D, Pagès M (2006) Maize DBF1-interactor protein 1 containing an R3H domain is a potential regulator of DBF1 activity in stress responses. Plant J 46:747–757
Shah K, Russinova E, Gadella TWJ Jr, Willemse J, de Vries SC (2002) The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev 16:1707–1720
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Sugikawa Y, Ebihara S, Tsuda K, Niwa Y, Yamazaki K (2005) Transcriptional coactivator MBF1s from Arabidopsis predominantly localize in nucleolus. J Plant Res 118:431–437
Takemaru K, Harashima S, Ueda H, Hirose S (1998) Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol Cell Biol 18:4971–4976
Tani T, Sobajima H, Okada K, Chujo T, Arimura S, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H (2008) Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227:517–526
Tsuda K, Yamazaki K (2004) Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim Biophys Acta 1680:1–10
Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14:903–916
Vermeer JE, Thole JM, Goedhart J, Nielsen E, Munnik T, Gadella TW Jr (2009) Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J 57:356–372
Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé DD, Veylder L (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15:59–63
von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12:452–457
Yoshioka S, Taniguchi F, Miura K, Inoue T, Yamano T, Fukuzawa H (2004) The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell 16:1466–1477
Zhu Y, Li Z, Xu B, Li H, Wang L, Dong A, Huang H (2008) Subcellular localizations of AS1 and AS2 suggest their common and distinct roles in plant development. J Integr Plant Biol 50:897–905
Acknowledgments
We are grateful to Dr Yasuo Niwa (University of Shizuoka) for kind providing pCaMV35S-sGFP(S65T)-nos3', Dr Hajime Yasui (Hokkaido University, Japan) for kindly providing the microscopes and to our colleagues for helpful discussions. This study was supported in part by a grant from the Sumitomo Foundation (to KM) and by Grants-in-Aid for the 21st COE (Center Of Excellence) Program ‘Marine Bio-Manipulation Frontier for Food Production’ and the City Area Program in Industry-Academia-Government Joint Research (Hakodate area) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to NS).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Uji, T., Takahashi, M., Saga, N. et al. Visualization of Nuclear Localization of Transcription Factors with Cyan and Green Fluorescent Proteins in the Red Alga Porphyra yezoensis . Mar Biotechnol 12, 150–159 (2010). https://doi.org/10.1007/s10126-009-9210-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-009-9210-5