Abstract
A two-parameter statistical model was used to predict the solubility of 96 putative virulence-associated proteins of Flavobacterium psychrophilum (CSF259-93) upon over expression in Escherichia coli. This analysis indicated that 88.5% of the F. psychrophilum proteins would be expressed as insoluble aggregates (inclusion bodies). These solubility predictions were verified experimentally by colony filtration blot for six different F. psychrophilum proteins. A comprehensive analysis of codon usage identified over a dozen codons that are used frequently in F. psychrophilum, but that are rarely used in E. coli. Expression of F. psychrophilum proteins in E. coli was often associated with production of minor molecular weight products, presumably because of the codon usage bias between these two organisms. Expression of recombinant protein in the presence of rare tRNA genes resulted in marginal improvements in the expressed products. Consequently, Vibrio parahaemolyticus was developed as an alternative expression host because its codon usage is similar to F. psychrophilum. A full-length recombinant F. psychrophilum hemolysin was successfully expressed and purified from V. parahaemolyticus in soluble form, whereas this protein was insoluble upon expression in E. coli. We show that V. parahaemolyticus can be used as an alternate heterologous expression system that can remedy challenges associated with expression and production of F. psychrophilum recombinant proteins.
Similar content being viewed by others
Introduction
Flavobacterium psychrophilum is the etiologic agent of bacterial cold water disease (BCWD) and rainbow trout fry syndrome (RTFS) in salmonid and other finfish. High mortality rates and spinal deformities associated with BCWD are responsible for serious economic losses to public and private aquaculture in many parts of the world (Nematollahi et al. 2003). Currently, there is no commercial vaccine available for the control of BCWD, and immunization of rainbow trout with whole-cell killed F. psychrophilum preparations have, in general, yielded limited success (Rahman et al. 2002). Protection against BCWD in rainbow trout can be induced after immunization with isolated antigenic fractions of F. psychrophilum (LaFrentz et al. 2004). Other F. psychrophilum antigens such as outer membrane proteins have also been shown to confer significant protection from experimental BCWD challenge (Rahman et al. 2002). Vaccine development studies have identified and characterized several immunogenic proteins, such as OmpH-like surface antigen p18 (Dumetz et al. 2006), Flavobacterium sp. protein antigen FspA (Crump et al. 2005), OmpA antigen (Dumetz et al. 2007), and Hsp60 and Hsp70 antigens (Sudheesh et al. 2007).
Identification of immunogenic proteins of F. psychrophilum has been primarily based on large-scale cultivation of F. psychrophilum followed by fractionation of antigens and testing their immunoreactivity to convalescent serum samples obtained from rainbow trout infected with F. psychrophilum. Because of the slow growth and fastidious nature of F. psychrophilum, this approach is time consuming and often allows identification of only those antigens that are abundant and can be purified in large quantities suitable for immunoprophylactic studies. Moreover, immunogenic proteins encoded by genes that are specifically induced during in vivo growth will be missed during such screening. Therefore, if the identified antigen is less abundant or induced specifically during infection, it needs to be produced on a large-scale by cloning and expression of the gene coding for that antigen. With the recent availability of whole genome sequences of F. psychrophilum strain JIP02/86 (Duchaud et al. 2007) and CSF259-93 (Wiens et al. unpublished data), it is now possible to use in silico analysis of genomic sequence to predict antigens that can be subsequently cloned and expressed in Escherichia coli for vaccine efficacy trials. This approach, in theory, could be used to produce many proteins in large quantity for testing in a fish challenge model. In this setting, E. coli is the preferred expression host because of the procedural simplicity, well known E. coli genetics, availability of compatible molecular tools, and relatively high yields per unit biomass; these traits can keep production costs relatively low. This also permits relatively simple construction and screening of high throughput expression libraries.
The elucidation of immunogenic, structural, or functional characteristics of the F. psychrophilum recombinant proteins is contingent on the availability of abundant recombinant proteins in soluble form. Expression of heterologous proteins in E. coli, however, can be limited by formation of insoluble aggregates called “inclusion bodies” (Sorensen and Mortensen 2005). Inclusion body proteins are devoid of biological activity and need elaborate solubilization, refolding, and purification procedures to recover products in their native conformation or in functionally active form (Rudolph and Lilie 1996; Vallejo and Rinas 2004). Moreover, loss of secondary structure during solubilization and the aggregation of denatured molecules because of intramolecular interactions during refolding result in poor recovery of proteins. Therefore, prior knowledge of F. psychrophilum sequences that do, or do not, tend toward soluble expression in E. coli is of particular relevance.
In this study, our purpose was to apply a statistical model (Wilkinson and Harrison 1991) to predict the solubility of putative virulence associated genes of F. psychrophilum CSF259-93 upon overexpression in E. coli. Experimental verification of solubility predictions of a limited set of proteins by colony filtration (CoFi) blot (Cornvik et al. 2005) showed that the expression of large molecular mass proteins (≥60 kDa) of F. psychrophilum in E. coli was associated with the formation of inclusion bodies. In addition, comprehensive analysis of differences in codon usage between F. psychrophilum and E. coli revealed a significant codon usage bias between these two organisms leading to reduced expression of full-length F. psychrophilum recombinant proteins in E. coli. Consequently, an alternative host expression system using V. parahaemolyticus was developed because of similar codon usage to that of F. psychrophilum. The effectiveness of this newly developed V. parahaemolyticus expression host system over E. coli was demonstrated after expression and purification in soluble form of a full-length recombinant F. psychrophilum hemolysin.
Materials and Methods
Database
DNA and protein sequences corresponding to 96 putative virulence-associated genes of F. psychrophilum were retrieved from the genome draft of F. psychrophilum CSF259-93 strain maintained at ERGO™ integrated genomics database (Wiens et al., unpublished data). These genes encode proteins with broad functional categories, such as bacterial outer membrane proteins, surface proteins, holin proteins, enterohemolysins, quorum-sensing-type proteins, S-layer toxin, and proteins encoding xanthan and alginate biosynthesis pathways. Domain lengths for these proteins ranged from 78 to 1,085 amino acid residues.
Solubility Predictions
In vivo solubility of each of the 96 F. psychrophilum proteins upon overexpression in E. coli was calculated by using Wikinson and Harrison’s (WH) two-parameter statistical model (Wilkinson and Harrison 1991; Davis et al. 1999). This solubility model is based on average charge as determined by the relative numbers of Asp, Glu, Lys, and Arg residues and the content of turn forming residues (Asn, Gly, Pro, and Ser). Briefly, the solubility prediction is based on a canonical value (CV), which is calculated as follows:\({\text{CV}} = \left[ {{\text{15}}{\text{.43}} \times \left( {N + G + P + S} \right) \div n} \right] - \left[ {\left( {{\text{29}}{\text{.56}} \times \left| {\left( {R + K} \right) - \left( {D + E} \right)} \right| \div n} \right) - {\text{0}}{\text{.03}}} \right]\) where N, G, P, S, R, K, D, and E represent the absolute number of Asn, Gly, Pro, Ser, Arg, Lys, Asp, and Glu residues in the protein, respectively, and n is the number of amino acids in the protein. If the difference between CV and CV′ (a discriminate whose value has been set to 1.71) is positive, the protein is predicted to be insoluble, and if the difference is negative, the protein is predicted to be soluble. The probability (P) of solubility was calculated as P = 0.4934 + 0.276 (CV − CV′) − 0.0392 (CV − CV′) (Harrison 2000). Using percentage probabilities to classify proteins as soluble or insoluble, a discriminant analysis has been used to successfully classify proteins as being soluble or insoluble with an overall accuracy of 88% (Harrison 2000).
E. coli Expression Constructs
Seven open reading frames (ORFs) of different lengths encoding putative virulence-associated proteins of F. psychrophilum CSF259-93 were selected based on global sequence features such as sequence length, isoelectric point (pI), grand hydropathicity index (GRAVY), aliphatic index (AI), and net charge and instability index (II), calculated using the ExPASy ProtParam tool (Gasteiger et al. 2003). The oligonucleotide primers used for amplification of complete ORFs are listed in Table 1.The complete ORFs were amplified using PfuUltra II fusion HS DNA polymerase (Stratagene, USA). The polymerase chain reaction (PCR)-amplified fragments were purified using QIAquick PCR Purification kit (Qiagen, USA) and ligated to pET101/D-TOPO and pET102/D-TOPO expression vectors (Invitrogen, USA) as per the protocol given by the manufacturer. pET101/D-TOPO allows expression of proteins with native N terminus and a C-terminal 6x-His tag, whereas pET102/D-TOPO is designed to express recombinant proteins with an additional N-terminal His-Patch thioredoxin (13 kDa) to facilitate optimum translation and, in some cases, increases the solubility of the recombinant proteins. The recombinant plasmids were transformed into one shot TOP10 chemically competent cells (Invitrogen), and positive recombinant clones were confirmed by PCR and DNA sequencing. The recombinant plasmids were then extracted using QIAprep Spin Miniprep kit (Qiagen) and stored at −20°C. An expression construct of ORF FpsyCSF259-93_0822, encoding a ∼60-kDa heat shock chaperonin of F. psychrophilum (Sudheesh et al. 2007), was amplified using primers 0822_F (5′-CACC ATGGCAAAAAATATAAAATTTG-3′) and 0822_R (5′-CATCATGCCTGGCATACC-3′) and cloned into pET102/D-TOPO as described above.
Experimental Determination of Solubility
Solubility of expressed protein was determined experimentally by a CoFi blot screening method (Cornvik et al. 2005). This method is based on separation of soluble protein from inclusion bodies by a filtration step at a colony level and has previously been used to screen expression constructs from a deletion mutagenesis library with 84% specificity for discrimination between soluble and insoluble expression products in the Rosetta (DE3) pLysS E. coli strain. Briefly, E. coli Rosetta2 cells (Novagen, USA) harboring the expression constructs were spot inoculated on two Luria–Bertani (LB) plates with 1% (w/v) glucose and grown overnight at 37°C. The plated colonies were then lifted with a 0.45-μm Durapore membrane (Millipore, USA), and the filter was placed with colonies facing up on LB plate containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Protein expression was induced for 5 h at 37°C (first set) and for 8 h at 30°C (second set). Subsequently, filters with colonies were placed on top of a 0.2-μm nitrocellulose filter and a Whatman 3MM paper soaked in native lysis buffer (20 mM Tris pH 8.0, 100 mM NaCl, 0.1 mg/ml lysozyme, 0.75 mg/ml DNAse I, 10 mM MgCl2, 1/2 Complete EDTA-free protease inhibitor cocktail tablet/10 ml [ethylenediaminetetraacetic acid (EDTA); Roche, USA]). The filter sandwich was incubated at room temperature for 30 min and then freeze–thawed three times with each cycle of 10 min at −80°C and 10 min at 37°C. The nitrocellulose membrane was removed from the sandwich, and the recombinant proteins bound to the membrane were detected with anti-His(C-term)-HRP antibody (Invitrogen) following the procedure described below. An expression construct of ORF FpsyCSF259-93_0822, encoding a soluble chaperonin, was used as a positive control for experimental determination of solubility.
Expression in E. coli
Immediately before expression of recombinant proteins, either BL21* (DE3) (Invitrogen) or Rosetta2 (Novagen) competent cells were transformed by heat shock method using ∼20 to 40 ng of purified recombinant plasmid. Subsequently, transformation mixtures were directly inoculated in LB medium containing 1% (w/v) glucose and grown overnight at 37°C, with shaking at 200 rpm. Next day, 10 ml of LB medium with no glucose was inoculated with 500 μl of overnight cultures and grown at 37°C with shaking at 200 rpm, until the OD595 of 0.5 to 1.0 was achieved. All constructs were then expressed by induction with 1 mM IPTG at 37°C for 4.5 to 5 h with shaking at 200 rpm. Unless specified, the E. coli BL21* (DE3) cells were grown in the presence of carbenicillin (100 μg/ml), and the Rosetta2 cells were grown in the presence of carbenicillin (100 μg/ml) and chloramphenicol (34 μg/ml). Cell pellets obtained from 1 ml of expression cultures were resuspended in phosphate-buffered saline (PBS; 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, 1 Complete EDTA-free protease inhibitor cocktail tablet/10 ml [Roche]) and sonicated for 1 min at level 10 with Misonix cup-horn sonicator (Misonix Inc., USA). The whole cell lysates were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by transfer to nitrocellulose membrane using Mini-Trans Blot cell (Bio-Rad, USA) as per the manufacturer’s instructions. Subsequently, the nitrocellulose membrane was removed from the sandwich and blocked with 5% skim milk in TBST (Tris-buffered saline Tween-20; 20 mM Tris–HCl, 500 mM NaCl, pH 7.5, 0.05% Tween 20) for 1 h. The membrane was washed 3 × 10 min with TBST and incubated for 1.5 h with anti-His(C-term)-HRP antibody (Invitrogen) diluted 1:5,000 in TBST. Next, the membrane was washed 3 × 10 min with TBST followed by 1 × 10 min with TBS and developed with the Immun-Star™ HRP Chemiluminescent kit (Bio-Rad) according to the manufacturer’s instructions.
Codon Usage Analysis of E. coli vs F. psychrophilum
Ninety-six genes, containing 48,290 codons, of F. psychrophilum CSF259-93 were used for codon usage analysis. The percent frequency of a codon is the frequency of occurrence of a given codon over the total number of codons in that gene. The percent frequency of 61 codons along with the position-specific mol% G + C at first, second, and third position “wobble base” codons was calculated using Codontree program (Pesole et al. 1988). The data obtained from the above analysis were used to calculate relative synonymous codon usage (RSCU) and relative adaptiveness (w) of each codon according to the previously described method (Sharp and Li 1987). An RSCU value for a codon is the observed frequency of the codon divided by its expected frequency under the assumption of equal usage of the synonymous codons for that amino acid. For relative adaptiveness calculations for each amino acid, the codon with the highest frequency was set to 100% relative adaptiveness, and all the other codons for the same amino acid were then scaled accordingly. For comparison, the RSCU values and relative adaptivenss of each of the 61 codons of expression host E. coli were calculated from the data retrieved from the codon usage a database maintained at the web server http://www.kazusa.or.jp/codon/. To predict the suitability of F. psychrophilum genes to the translational system of E. coli, codon adaptation index (CAI) values for F. psychrophilum genes were calculated as described by Sharp and Li (1987) using the Java Codon adaptation tool (JCat) program (Grote et al. 2005). CAI is the geometric mean of the RSCU values corresponding to each of the codons used in that gene divided by the maximum possible CAI for a gene of the same amino acid composition. Usually, CAI is estimated by examining a set of highly expressed reference genes, and, thus, it is an estimator of gene expressivity through codon usage.
In silico Optimization of Codon Usage in F. psychrophilum Genes for Adaptation in E. coli Expression Host
In silico codon optimization of eight F. psychrophilum genes for their adaptation in an E. coli expression host was performed using JCat program (Grote et al. 2005). This novel tool allows the codon optimization of a target gene such that rare codons in heterologous genes are replaced with most frequently used codons by the expression host. The synthetically designed gene is then well adapted to the codon usage of its potential expression host. The sequence homology between eight E. coli-codon optimized gene sequences with that of native gene sequences was determined using ClustlW program at the web server http://www.ebi.ac.uk/Tools/webservices.
Cloning of F. psychrophilum Putative Hemolysin for Expression in Alternative Host V. parahaemolyticus
The complete ORF of F. psychrophilum putative hemolysin (FpsyCSF259-93_0066) was amplified using a forward primer 0066-PMMB-F 5′-GAGCTCgaaggagaTATACATATGGGTTTAGTTACCGCTAA-3′ (Sac1 restriction site is indicated in italics, small letters indicate a ribosomal binding site that is followed by a spacer sequence and an underlined start codon) and a reverse primer 0066-PMMB-R 5′-GGTACC TTAatgatgatgatgatgatgGCACTCCCCTTTTTCTGCTA-3′ (Kpn1 restriction site is indicated in italics, underlined letters indicate stop codon, and small letters indicate 6x-His tag that will be incorporated at C-terminal of an expressed product) according to the procedure described above. The amplified PCR product was ligated to pCR4.0 vector (Invitrogen) according to the protocol given by the manufacturer. The recombinant pCR4.0 plasmid was then digested with Sac1 and Kpn1 enzymes, and the digested fragment of ORF0066 with flanking Sac1–Kpn1 restriction sites was purified using QiaxII gel purification kit (Qiagen) as per the manufacturer’s instructions. The restriction fragment was ligated to pMMB207 plasmid predigested with Sac1 and Kpn1 (Morales et al. 1991). The expression of heterologous genes in pMMB207 is driven by IPTG inducible tac promoter. Because pMMB207 plasmid does not have a start codon at a suitable distance after a ribosome-binding site (RBS), an RBS followed appropriately by a start codon was incorporated at 5′ end of the forward primer. The resultant recombinant plasmid 1390-pMMB207 was transformed to E. coli S17 λpir by electroporation. The recombinant clones were selected for chloramphenicol resistance encoded on pMMB207 and confirmed by PCR amplification of hemolysin gene.
V. parahaemolyticus NY-4 Ampr strain was transformed by conjugal transfer of 1390-pMMB207. Briefly, V. parahaemolyticus NY-4 Ampr strain was grown overnight in LB medium containing 2.5% (w/v) NaCl (LB salt) and ampicillin (100 μg/ml), at 37°C with shaking at 200 rpm. Recombinant E. coli S17 λpir strain containing 1390-pMMB207 was grown overnight in LB medium containing chloramphenicol (34 μg/ml), at 37°C with shaking at 200 rpm. After overnight incubation, 1 ml of each culture was centrifuged, washed once in LB salt, and resuspended in 50 μl of LB salt. The resuspended cells were mixed and spot inoculated on LB agar containing 2.5% NaCl followed by overnight incubation at 37°C. To select transconjugants, the mating mixture was plated on LB agar plates containing 2.5% NaCl, ampicillin, and chloramphenicol. The resultant transconjugants were confirmed by PCR amplification of the hemolysin gene and stored in 15% (v/v) glycerol at −80°C until further use.
Expression and Solubility Determination of Recombinant Hemolysin from V. parahaemolyticus NY-4 Strain
For the expression of recombinant hemolysin, V. parahaemolyticus NY-4 strain containing 1390-pMMB207 was grown in LB salt containing appropriate antibiotics until an OD600 of 0.5 to 1.0 was achieved. The expression was then induced by addition of 0.5 mM IPTG, and the culture was grown for another 5 h at 37°C with shaking at 200 rpm. The whole-cell pellet obtained from 1 ml of expression culture was resuspended in PBS, sonicated, and analyzed by Western blot as described above. The solubility of recombinant hemolysin expressed in V. parahaemolyticus was determined by CoFi blot as described for E. coli.
Purification of Soluble and Insoluble Recombinant Hemolysin
E. coli BL21* (DE3), Rosetta2, and V. parahaemolyticus NY-4 containing recombinant hemolysin constructs were grown in a 50-ml LB medium supplemented with appropriate antibiotics. Expression was induced with IPTG as described previously. Expression cultures were harvested by centrifugation at 4,000 rpm for 20 min. For the purification of soluble hemolysin, cells were resuspended and lysed in a native lysis buffer (50 mM Na2HPO4, 500 mM NaCl, 10 mM imidazole, pH 8.0, 80 mg lysozyme/10 ml, 50 μg DNase-I/10 ml, 10 mM MgCl2, and 1 Complete EDTA-free protease inhibitor cocktail tablet/10 ml), sonicated and centrifuged. Supernatants were loaded onto NiNTA agarose column (Invitrogen) following the manufacturer’s protocol for purification under native conditions. The columns were washed three times with a lysis buffer (50 mM Na2HPO4, 500 mM NaCl, 20 mM imidazole, pH 8.0), and elusion was performed with a lysis buffer containing 500 mM imidazole. For purification of insoluble hemolysin, cells were resuspended and lysed in a denaturing lysis buffer (20 mM Na2HPO4, 500 mM NaCl, 8 M urea, pH 8.0), sonicated and centrifuged. Supernatants were loaded onto NiNTA agarose column (Invitrogen) following the manufacturer’s protocol for purification under denaturing conditions. The columns were washed with a denaturing lysis buffer at pH 6.0 followed by two washes at pH 5.3, and elusion was performed by using a denaturing lysis buffer at pH 4.0. All proteins were checked by SDS-PAGE and Western blot analysis as described above. The quantity of total protein from crude lysate and from column purified protein was estimated by using a Micro BCA Protein Assay Reagent kit (Pierce, USA).
Results
Solubility Predictions
Analysis of 96 F. psychrophilum virulence-associated proteins according to WH method showed that 11 (11.5%) proteins were predicted to be soluble in E. coli, with a CV − CV′ ranging from −0.09 to −5.15 (Fig. 1) and the probabilities of solubility ranging from 51.8% to 97%, respectively. On the other extreme, 85 (88.5%) F. psychrophilum proteins were predicted to be insoluble in E. coli, with a CV − CV′ ranging from 3.04 to 0.01 (Fig. 1) and probabilities of solubility ranging from 3.4% to 50.3%, respectively.
Distribution of CV − CV′ values for 96 F. psychrophilum virulence associated proteins. CV − CV′ values computed for all proteins were plotted against the length of amino acid sequence. Negative CV − CV′ values (plotted as dark black circles) predict the protein to be soluble, while the positive values (plotted as triangles) predict the protein to be insoluble on expression in E. coli
Expression in E. coli
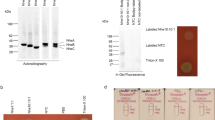
The global sequence features including solubility predictions of F. psychrophilum proteins selected for experimental expression in E. coli are shown in Table 2. The molecular size of selected proteins ranged from ∼60 to ∼101.8 kDa. These were selected because of the recent evidence suggesting that high molecular mass antigens of F. psychrophilum are immunogenic and provide significant levels of protection against experimental BCWD challenge of rainbow traut (LaFrentz et al. 2004). Initially, seven ORFs (Table 1) were cloned in pET101/D-TOPO vector and expressed in E. coli BL21* (DE3) cells. Most of the pET101 constructs did not express proteins in sufficient quantities for reliable detection by SDS-PAGE or Western blot (data not shown). Consequently, all seven ORFs were cloned into pET102/D-TOPO vector, co-expressing a 13-kDa N-terminal thioredoxin fusion tag, and expressed in E. coli BL21* (DE3) cells. For five out of seven pET102 constructs (FpsyCSF259-93_1419, 2309, 0896, 0066, and 0586), we were able to detect proteins by Western blot when probed with anti-His(C-term)-HRP antibody (Fig. 2a). The above five expression constructs consistently expressed full-length proteins of expected sizes (small black arrows, Fig. 2) together with one or more minor molecular weight species. Expression of FpsyCSF259-93_2104 and FpsyCSF259-93_0706 was inconsistent and hence excluded from further analysis. We also expressed all five expression constructs in Rosetta2 host strain, which is genetically engineered to co-express tRNAs for the seven codons (ATA, AGG, AGA, CUA, CCC, GGA, and CGG) rarely utilized in E. coli. At least three (AGA, GGA, and ATA) of the seven rare codons were identified as problematic in F. psychrophilum genes. Co-expression of rare tRNA genes led to the reduction in the numbers of extra minor molecular weight species in most expression constructs; however, these problems were not completely alleviated using this approach (Fig. 2b).
Western blot analysis of whole cell lysates of a E. coli BL21* (DE3) and b Rosetta2 host strains showing expression of full-length proteins of F. psychrophilum (indicated with small black arrows) along with minor molecular weight species: myosin cross-reactive antigen (lane 1, ∼88 kDa), OMP bacterial surface protein (lane 2, ∼78 kDa), OmpA/MotB family protein (lane 3, ∼94 kDa), hemolysin (lane 4, ∼85 kDa), virulence-associated protein E (lane 5, ∼96 kDa), and GroEL heat shock protein (lane 6, ∼76 kDa). c Shows Western blot detection of recombinant F. psychrophilum hemolysin expressed and purified from E. coli BL21 (lane 1), E. coli Rosetta2 (lane 2), and V. parahaemolyticus (lane 3). Note that the vector used for expression in V. parahaemolyticus did not include N-terminal thioredoxin fusion and is therefore 13-kDa smaller than expected product sizes from E. coli. M precision plus protein standard (Bio-Rad)
Experimental Verification of Solubility
Experimental verification of solubility of recombinant proteins produced from five expression constructs (FpsyCSF259-93_1419, 2309, 0896, 0066, and 0586) by CoFi blot showed no or very limited signal intensities when expressed at 37°C (Fig. 3), indicating that the proteins were expressed in the form of insoluble inclusion bodies (see below). Expression at reduced growth temperature (30°C) did not have any positive effects on the solubility of recombinant proteins as evident by the extremely low signal intensities on the CoFi blots (Fig. 3). As expected, the CoFi blot of recombinant protein expressed from FpsyCSF259-93_0822 showed intense signal, indicating that this protein was expressed in the soluble form.
Results of colony filtration blot (CoFi) for the identification of soluble protein expression in E. coli Rosetta2 (i) and V. parahaemolyticus (ii). (i) E. coli colonies expressing F. psychrophilum proteins: myosin cross-reactive antigen (lane 1), OMP bacterial surface protein (lane 2), OmpA/MotB family protein (lane 3), hemolysin (lane 4), virulence-associated protein E (lane 5), GroEL heat shock protein (lane 6), and (ii) V. parahaemolyticus colony-expressing F. psychrophilum hemolysin were induced at 37°C (a) or 30°C (b) and lysed on membrane filter. Soluble protein from each colony diffuses through the filter and is captured on nitrocellulose membrane placed under the membrane filter. Detection with anti-His(C-term)-antibody shows spots indicating presence or absence of soluble protein. Intensity of spot corresponds to the yield of soluble protein
Codon Usage Analysis of E. coli vs F. psychrophilum
Overall, the position-specific mol %G + C for codons in F. psychrophilum genes was 40.7% at the first position, 34% at the second position, and 24% at the third position. In contrast, %G + C at the first, second, and third positions of codons in E. coli was 58.9%, 40.6%, and 55%, respectively. For an index of codon usage bias, RSCU values for F. psychrophilum genes and E. coli were calculated with the percent frequency values of codon usage in F. psychrophilum and E. coli, respectively (Table 3). The selection of optimal codons was based on the values used for E. coli. An RSCU value ≥ 1 indicates that the codon is used optimally or more frequently than expected in a given organism. Similarly, an RSCU value < 1 means that the codon is used, but at a rate lower than expected. Examination of codon usage based on RSCU values allowed selection of ten codons (underlined letters in Table 3) with RSCU values greater than 1 in both F. psychrophilum and E. coli, indicating that these codons are utilized equally well in both the organisms. Sixteen codons were identified with RSCU value < 1 in both F. psychrophilum as well as E. coli, indicating that these codons were used rarely or at lower rates in these organisms. In contrast, 15 unique codons (asterisk in Table 3) with RSCU values > 1 were identified in expression host E. coli, signifying that these were frequently used in E. coli, but rarely used in F. psychrophilum. Interestingly, 18 unique codons were identified with RSCU values of >1 in F. psychrophilum but <1 in E. coli (bold faced letters in Table 3), indicating that these codons are frequently used in F. psychrophilum, but rarely in E. coli.
Eighteen frequently used codons in F. psychrophilum reflected bias of A or T over G or C at third position “wobble” base (17 codons end with A or T, while only 1 codon ends with G, and none with C). Examination of the distribution of these 18 codons varied markedly between the seven ORFs that were cloned for this study (data not shown). Consequently, the CAI for all eight F. psychrophilum genes was calculated to assess their level of codon adaptability and expressibility in E. coli (Table 4). CAI values for F. psychrophilum genes ranged from 0.159 to 0.238, suggesting poor codon adaptability and lower expressibility of these genes in E. coli. We then employed the JCat program to “design” F. psychrophilum genes that would be optimized for expression in E. coli (CAI = 1). The %G + C content of native F. psychrophilum genes ranged between 29.7% and 39.2%, whereas in the E. coli-codon optimized genes, %G + C content increased by 9.1% to 15.6%. Overall, the codon optimized and the native F. psychrophilum gene sequences showed 73% to 79% sequence similarity.
V. parahaemolyticus as an Alternative Expression Host
Synthetic production of E. coli optimized genes would be justified if the genes in question were known to be suitable candidates for further study. Lacking this information, a more parsimonious course of action would be to identify an alternative expression host. In this study, we evaluated V. parahaemolyticus as an alternative expression host for the optimal expression of recombinant F. psychrophilum proteins. Codon usage analysis of V. parahaemolyticus showed that, with the exception of three codons (AGA, GGA, and ATA), the relative adaptiveness of most of the other codons closely matched between V. parahaemolyticus and F. psychrophilum (Table 3); thus, V. parahaemolyticus has a potential to express recombinant F. psychrophilum proteins much more efficiently than E. coli. To empirically assess this conclusion, we cloned and expressed F. psychrophilum hemolysin in V. parahaemolyticus (strain NY-4) and compared the quality and quantity of expressed protein with that of similar protein expressed from E. coli BL21* (DE3) and Rosetta2 hosts (Table 5). A recombinant hemolysin with >90% purity was recovered from E. coli BL21* (DE3), Rosetta2, and V. parahaemolyticus, respectively, and constituted 5.2%, 4%, and 3.5 to 4.7% of the total protein obtained from expression cultures of the respective strains (Table 5). As expected, E. coli produced previously observed lower molecular weight products, but the purified extract from V. parahaemolyticus yielded protein of expected size without any lower molecular weight products (Fig. 2c). The recombinant hemolysin could be extracted from E. coli only when denaturing conditions were employed to solubilize protein from inclusion bodies (Fig. 4). In contrast, recombinant hemolysin from V. parahaemolyticus was extracted using both native (non-denaturing) as well as denaturing conditions. Furthermore, a CoFi blot showed that the recombinant hemolysin was present in a soluble form when overexpressed in V. parahaemolyticus and that the solubility was improved when expression was induced at 30°C (Fig. 3).
SDS-PAGE showing solubility of F. psychrophilum recombinant hemolysin upon overexpression in E. coli BL21 (lanes 1 and 2), Rosetta2 (lanes 3 and 4), and V. parahaemolyticus (lanes 5 and 6). The protein expression was induced at 37°C for 5 h using 1 mM (for E. coli) and 0.5 mM (for V. parahaemolyticus) IPTG. The expressed proteins were purified from the cell pellets of expression cultures by binding of 6x-His tag using Ni2+-NTA matrices using native conditions (lanes 1, 3, and 5) and denaturing conditions using 8 M urea (lanes 2, 4, and 6). M broad range protein standard (Bio-Rad)
Discussion
Recombinant expression of F. psychrophilum genes represents the simplest approach for obtaining abundant protein for further studies. Characterization of these proteins, however, is contingent upon availability of abundant and soluble material. According to the WH method (Wilkinson and Harrison 1991; Davis et al. 1999), the majority (88.5%) of F. psychrophilum proteins will form insoluble aggregates (inclusion bodies) upon expression in E. coli, indicating that the effective recovery of these proteins will require elaborate solubilization, refolding, and purification procedures. Our solubility predictions were confirmed by experimental expression of a set of F. psychrophilum proteins with a range of solubility probabilities and different global sequence features. These proteins were co-expressed with the 13-kDa N-terminal thoredoxin fusion tag and subjected to a fairly simple but robust technique of CoFi blot to document solubility (Cornvik et al. 2005). As expected, the signal intensity obtained from the colony expressing the ∼60-kDa F. psychrophilum chaperonin (FpsyCSF259-93_0822) protein was high, indicating higher yield of soluble protein. This outcome was predicted given a CV − CV′ of −0.17. In contrast, the CoFi blot for recombinant proteins expressed from five other expression constructs (FpsyCSF259-93_1419, 2309, 0896, 0066, and 0586) showed little signal indicating that all of these proteins were expressed in an insoluble form. Recombinant protein expression at reduced growth temperatures can increase the solubility of aggregation prone proteins (Schein and Noteborn 1988), but our efforts to express F. psychrophilum proteins in E. coli at reduced growth temperatures (30°C for 8 h) did not improve solubility. Interestingly, when the recombinant proteins were expressed in the E. coli BL21* (DE3), Western blots indicated presence of full-length proteins from all the five ORFs (FpsyCSF259-93_1419, 2309, 0896, 0066, and 0586), but these products were consistently associated with one or more minor molecular weight products. We hypothesized that these minor molecular weight products are a result of translational errors because of the differences in codon usage in F. psychrophilum genes.
Expression of heterologous genes encoded by codons that are rarely used by E. coli can lead to translational errors such as mistranslational amino acid substitution, frame shifting events, or premature termination of translation (Kurland and Gallant 1996; McNulty et al. 2003; Sorensen et al. 2003). It is less likely that the minor molecular weight products observed in this study are truncated species because of the premature termination of translation or +1 frame shift events because the 6x-His tag is located at C terminus of protein and the Western blot with anti-His antibody would only detect full-length protein products. It is possible, however, that the depletion of intracellular pool of cognate tRNAs may have led to ribosomal pause at codons rarely used by E. coli thereby causing in-frame deletion of amino acids (because of hopping over of ribosome to next optimally used codon) leading to production of minor molecular weight products that are translated in-frame, but reduced in size (Kane et al. 1992; Wysocki et al. 1994; Kane 1995). In addition, rare codon-mediated ribosomal stalling within the mRNA transcript can also alter the RNA structure by generating an internal ribosomal entry site and facilitate translation of the downstream ORF in the presence of internal initiation codons (Narayanan and Dubnau 1987; Fernandez et al. 2005). This can lead to production of in-frame translated minor molecular weight products. We suspect this occurred in our case because all of the ORFs expressed in this study showed presence of several in-frame internal initiation codons and, in some cases, Shine–Dalgarno like sequences (data not shown). Thus, some of the minor molecular weight species observed in this study could be attributed to combined effects of ribosomal stalling because of rare codons and initiation of translation from internal initiation codons. Sequencing analysis of expressed protein products would be required to further confirm this effect; however, we did not investigate this further, as this was beyond the scope of our study.
We attempted to alleviate these problems by co-expressing rare tRNAs to better match codon usage in our cloned F. psychrophilum sequences. The Rosetta2 strain used in this study is engineered to co-express seven most commonly reported problematic tRNA genes (ATA, AGG, AGA, CTA, CCC, GGA, and CGG) of which at least three codons (ATA, GGA and AGA) were identified to be problematic in F. psychrophilum genes. As expected, this strategy could only partially remedy the problem, as evident by the reduction in the numbers of extra minor molecular weight species for most expression constructs. Nevertheless, these results indicated that the production of minor molecular weight species were associated with the codon usage bias; computation analysis of 96 F. psychrophilum ORFs was consistent with a pervasive codon bias problem when attempting to express these genes in an E. coli host. Others have reported that the overall G + C composition is a major factor affecting codon use variation, and as a result, increased frequency of rare codon usage can markedly affect quality and quantity of expression (Kane, 1995; Chen et al. 2004). Noteworthy is that the mol %G + C between E. coli (∼51%) and F. psychrophilum (∼32%) is significantly different, and we also observed that F. psychrophilum codon usage is biased toward codons ending in A or T over codons ending in G or C. Taken together, our analysis indicates that the codon preference in these organisms is strikingly different and that the numbers of tRNA genes co-expressed in this study were not sufficient to alleviate problems of codon usage bias.
The lower CAI values (0.159–0.238) for eight F. psychrophilum ORFs indicated that, if F. psychrophilum proteins are expressed in E. coli, their expression would be suboptimal (Sharp and Li 1987). The CAI analysis also indicated that, for efficient expression of F. psychrophilum genes, codon usage must be optimized to match the expression host E. coli. Problems of mistranslation and reduced expression levels can be alleviated when rare codons are substituted with the optimal codons (Kane et al. 1992; Calderone et al. 1996). Codon optimization can improve gene expression through different mechanisms, such as improvement in translation by avoiding the requirement for tRNAs present at low concentrations and increase in mRNA stability with a rise in overall G + C content (Narum et al. 2001). Therefore, it is likely that synthesizing new F. psychrophilum genes to include more appropriate codons would improve the expression and our in silico analysis indicates that this strategy is feasible. Indeed, it is now possible to rapidly synthesize even large gene sequences (approximately 20 kb) in a relatively short time (<4 weeks/kb). However, this approach could be cost limiting especially when attempting to characterize the function and immunogenic properties of a large number of proteins.
A more parsimonious solution to codon bias limitations is to develop new heterologous expression systems that better match codon use patterns of the target organism. This strategy is becoming increasingly popular, and several bacterial hosts, such as Bacillus megaterium, B. subtilis, B. brevis, and Caulobacter crecentus, have been described for heterologous protein expression (Terpe 2006). Our comparison of codon usage between V. parahaemolyticus with that of F. psychrophilum showed that, with the exception of three codons (AGA, GGA, and ATA), usage is very similar. Moreover, genetic tools are also available for the recombinant protein expression in V. parahaemolyticus. These features of V. parahaemolyticus aided expression of a full-length hemolysin (∼69 kDa) protein, which was produced without any minor molecular mass products. In addition, the recombinant hemolysin was expressed in a soluble form in V. parahaemolyticus, which is in sharp contrast to E. coli where the same hemolysin is expressed in the form of insoluble inclusion body aggregates. This is important because purification of soluble hemolysin produced in V. parahaemolyticus did not require elaborate solubilization using high concentrations of chaotropic agents, and hence, tedious and time-consuming post-purification processing steps were avoided (e.g., dialysis to remove chaotropic agents, refolding, and concentration of dialyzed protein). Finally, the quantity of purified recombinant hemolysin produced in V. parahaemolyticus was comparable to that of E. coli. Collectively, these results indicate that V. parahaemolyticus can be used for the overexpression and production of recombinant F. psychrophilum proteins.
In conclusion, overexpression of large molecular weight proteins from F. psychrophilum in the E. coli host is associated with recombinant protein insolubility; codon bias leads to poor quality proteins. These problems are fundamental constraints for production of proteins needed for immunoprophylactic and structural or functional studies. Our study provides a solution to these problems in the form of V. parahaemolyticus as an alternative expression host that is suitable for production of high-quality and soluble recombinant protein.
References
Calderone TL, Stevens RD, Oas TG (1996) High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J Mol Biol 262:407–412
Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH (2004) Codon usage between genomes is constrained by genome-wide mutational processes. Proceedings of the National Academy of Sciences of the United States of America 101:3480–3485
Cornvik T, Dahlroth SL, Magnusdottir A, Herman MD, Knaust R, Ekberg M, Nordlund P (2005) Colony filtration blot: a new screening method for soluble protein expression in Escherichia coli. Nat Methods 2:507–509
Crump EM, Burian J, Allen PD, Kay WW (2005) Identification and expression of a host-recognized antigen, FspA, from Flavobacterium psychrophilum. Microbiology 151:3127–3135
Davis GD, Elisee C, Newham DM, Harrison RG (1999) New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol Bioeng 65:382–388
Duchaud E, Boussaha M, Loux V, Bernardet JF, Michel C, Kerouault B, Mondot S, Nicolas P, Bossy R, Caron C, Bessieres P, Gibrat JF, Claverol S, Dumetz F, Le Henaff M, Benmansour A (2007) Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat Biotechnol 25:763–769
Dumetz F, Duchaud E, LaPatra SE, Le Marrec C, Claverol S, Urdaci MC, Le Henaff M (2006) A protective immune response is generated in rainbow trout by an OmpH-like surface antigen (P18) of Flavobacterium psychrophilum. Appl Environ Microbiol 72:4845–4852
Dumetz F, Lapatra SE, Duchaud E, Claverol S, Le Henaff M (2007) The Flavobacterium psychrophilum OmpA, an outer membrane glycoprotein, induces a humoral response in rainbow trout. J Appl Microbiol 103:1461–1470
Fernandez J, Yaman I, Huang C, Liu H, Lopez AB, Komar AA, Caprara MG, Merrick WC, Snider MD, Kaufman RJ, Lamers WH, Hatzoglou M (2005) Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol Cell 17:405–416
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, Jahn D (2005) JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33:W526–531
Harrison RG (2000) Expression of soluble heterologous proteins in Escherichia coli. inNovations 11:4–7
Kane JF (1995) Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol 6:494–500
Kane JF, Violand BN, Curran DF, Staten NR, Duffin KL, Bogosian G (1992) Novel in-frame two codon translational hop during synthesis of bovine placental lactogen in a recombinant strain of Escherichia coli. Nucleic Acids Res 20:6707–6712
Kurland C, Gallant J (1996) Errors of heterologous protein expression. Curr Opin Biotechnol 7:489–493
LaFrentz BR, LaPatra SE, Jones GR, Cain KD (2004) Protective immunity in rainbow trout Oncorhynchus mykiss following immunization with distinct molecular mass fractions isolated from Flavobacterium psychrophilum. Dis Aquat Organ 59:17–26
McNulty DE, Claffee BA, Huddleston MJ, Porter ML, Cavnar KM, Kane JF (2003) Mistranslational errors associated with the rare arginine codon CGG in Escherichia coli. Protein Expr Purif 27:365–374
Morales VM, Backman A, Bagdasarian M (1991) A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47
Narayanan CS, Dubnau D (1987) Demonstration of erythromycin-dependent stalling of ribosomes on the ermC leader transcript. J Biol Chem 262:1766–1771
Narum DL, Kumar S, Rogers WO, Fuhrmann SR, Liang H, Oakley M, Taye A, Sim BK, Hoffman SL (2001) Codon optimization of gene fragments encoding Plasmodium falciparum merzoite proteins enhances DNA vaccine protein expression and immunogenicity in mice. Infect Immun 69:7250–7253
Nematollahi A, Decostere A, Pasmans F, Haesebrouck F (2003) Flavobacterium psychrophilum infections in salmonid fish. J Fish Dis 26:563–574
Pesole G, Attimonelli M, Liuni S (1988) A backtranslation method based on codon usage strategy. Nucleic Acids Res 16:1715–1728
Rahman H, Kuroda A, Dijkstra JM, Kiryu I, Nakanishi T, Ototake M (2002) The outer membrane fraction of Flavobacterium psychrophilum induces protective immunity in rainbow trout and ayu. Fish Shellfish Immunol 12:169–179
Rudolph R, Lilie H (1996) In vitro folding of inclusion body proteins. Faseb J 10:49–56
Schein CH, Noteborn MHM (1988) Formation of soluble recombinant proteins in Escherichia coli is favored by lower growth temperatures. Bio/Technology 6:291–294
Sharp PM, Li WH (1987) The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol 4:222–230
Sorensen HP, Mortensen KK (2005) Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol 115:113–128
Sorensen HP, Sperling-Petersen HU, Mortensen KK (2003) Production of recombinant thermostable proteins expressed in Escherichia coli: completion of protein synthesis is the bottleneck. J Chromatogr B Analyt Technol Biomed Life Sci 786:207–214
Sudheesh PS, LaFrentz BR, Call DR, Siems WF, LaPatra SE, Wiens GD, Cain KD (2007) Identification of potential vaccine target antigens by immunoproteomic analysis of a virulent and a non-virulent strain of the fish pathogen Flavobacterium psychrophilum. Dis Aquat Organ 74:37–47
Terpe K (2006) Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 72:211–222
Vallejo LF, Rinas U (2004) Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microb Cell Fact 3:11
Wilkinson DL, Harrison RG (1991) Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology (N Y) 9:443–448
Wysocki LA, Myers DP, Kane JF (1994) A multiplex-PCR for the differentiation of Mycobacterium bovis and Mycobacterium tuberculosis. Faseb J 8(suppl 7):A1305
Acknowledgments
We thank S. Dobry, S. LaFrentz, and L. Orfe for their skillful technical assistance at various stages of this work. Initial version of FsyCSF259-93_0822 clone was provided by Karen Plant, Hagerman Fish Culture Experiment Station, Hagerman, ID, USA. This project was funded in part by WSU-UOI Aquaculture Initiative and by the Agricultural Animal Health Program at the College of Veterinary Medicine, Washington State University, Pullman, WA, USA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shah, D.H., Cain, K.D., Wiens, G.D. et al. Challenges Associated with Heterologous Expression of Flavobacterium psychrophilum Proteins in Escherichia coli . Mar Biotechnol 10, 719–730 (2008). https://doi.org/10.1007/s10126-008-9111-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-008-9111-z