Abstract
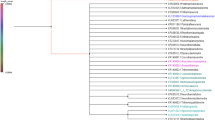
Genetic variation and population structure of hair crab (Erimacrus isenbeckii) were examined using nucleotide sequence analysis of 580 base pairs (bp) in the 3′ portion of the mitochondrial cytochrome c oxidase subunit I gene (COI) of 20 samples collected from 16 locales in Japan (the Hokkaido and Honshu Islands) and one in Korea. A total of 27 haplotypes was defined by 23 variable nucleotide sites in the examined COI region. Pairwise population F ST estimates and neighbor-joining tree inferred distinct genetic differentiation between the representative samples from the Pacific Ocean off the Eastern Hokkaido Island and the Sea of Japan, while others were intermediate between these two groups. AMOVA also showed a weak but significant differentiation among these three groups. The present results suggest a moderate population structure of hair crab, probably influenced by high gene flow between regional populations due to sea current dependent larval dispersal of this species.



Similar content being viewed by others
References
Azuma N, Kunihiro Y, Sasaki J, Nozawa Y, Mihara E, Mihara M, Yasunaga, Abe S (2006) Genetic variation of hair crab (Erimacrus isenbeckii) inferred from mitochondrial DNA sequence analysis. Fish Genet Breed Sci 35, 35–42
Baldwin JD, Bass AL, Bowen BW, Clark WC (1998) Molecular phylogeny and biogeography of the marine shrimp Penaeus. Mol Phylogenet Evol 10, 399–407
Bunch T, Highsmith RC, Shields GF (1998) Genetic evidence for dispersal of larvae of Tanner crabs (Chionoecetes bairdi) by the Alaskan Coastal Current. Mol Mar Biol Biotechnol 7, 153–159
Cassone BJ, Boulding EG (2006) Genetic structure and phylogeography of the lined shore crab, Pachygrapsus crassipes, along the northeastern and western Pacific coasts. Mar Biol 149, 213–226
Chow S, Abe S, Ueno Y, Torisawa M, Fujio Y (1987) Homozygote excess observed in natural population of horsehair crab Erimacrus isenbeckii. Nippon Suisan Gakkaishi 53, 321
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9, 1657–1659
Creasey S, Rogers A, Tyler P, Young C, Gage J (1997) The population biology and genetics of the deep-sea spider crab, Encephaloides armstrongi Wood-Mason 1891 (Decapoda: Majidae). Philos Trans R Soc Lond 352, 356–376
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular valiance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491
Felsenstein J (1993) PHYLIP (Phylogeny Interference Package) Version 3.5c. Department of Genetics, Washington State University Seattle
Geller JB, Walton ED, Grosholz ED, Ruiz GM (1997) Cryptic invasions of the crab Carcinus detected by molecular phylogeography. Mol Ecol 6, 901–906
Gopurenko D, Hughes JM, Keenan CP (1999) Mitochondrial DNA evidence for rapid colonisation of the Indo-West Pacific by the. mud crab Scylla serrata. Mar Biol 134, 227–233
Herborg LM, Weetman DW, Oosterhout C, Hänfling B (2007) Genetic population structure and contemporary dispersal patterns of a recent European invader, the Chinese mitten crab, Eriocheir sinensis. Mol Ecol 16, 231–242
Imai H, Cheng JH, Hamasaki K, Numachi K (2004) Identification of four mud crab species (genus Scylla) using ITS-1 and 16S rDNA markers. Aquat Liv Res 17, 31–34
Kawai H (1972) “Hydrography of the Kuroshio Extension”. In: Kuroshio, Stommel H, Yoshida K eds. (Tokyo: University of Tokyo Press) pp 235–352
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions that compare studies of nucleotidesequences. J Mol Evol 16, 111–120
Knowlton N, Weight LA (1998) New dates and new rates for divergence across the Isthmus of Panama. Proc R Soc Lond B 265, 2257–2263
Lessios HA, Kessing BD, Pearse JS (2001) Population structure and speciation in tropical seas: global phylogeography of the sea urchin diadema. Evolution 55, 955–975
Lessios HA, Kane J, Robertson DR (2003) Phylogeography of the pantropical sea urchin Tripneustes: contrasting patterns of population structure between oceans. Evolution 57, 2026–2036
McMillen-Jackson AM, Bert TM, Steele P (1994) Population genetics of the blue crab Callinectes sapidus: modest population structuring in a background of high gene flow. Mar Biol 118, 53–65
Mihara E (2003) “Kegani Erimacrus isenbecckii (Brandt)”. In: Gyogyou Seibutu Zukan, Shin Kita no Sakanatachi, Mizushima T, Torisawa M eds. (Sapporo, Japan: Hokkaido Shinbun sha) pp 380–385 (in Japanese)
Naganuma K (1977) The oceanographic fluctuations in the Japan Sea. Mar Sci (Kaiyo Kagaku) 9, 137–141 (in Japanese with English abstract)
Nagashima K, Sato M, Kawamata K, Nakamura A, Ohta T (2005) Genetic structure of Japanese scallops population in Hokkaido, analyzed by mitochondrial haplotype distribution. Mar Biotechnol 7, 1–10
Nei M (1987) Molecular Evolutionary Genetics (New York: Columbia University Press)
Nei M, Tajima F (1981) DNA polymorphism detectable by restriction endonucleases. Genetics 97, 145–163
Palumbi SR (1994) Genetic divergence, reproductive isolation, and marine speciation. Rev Ecol Syst 25, 547–572
Pfeiler E, Hurtado LA, Knowles LL, Torre-Cosío J, Bourillón-Moreno L, Márquez-Farías JF, Montemayor-López G (2005) Population genetics of the swimming crab Callinectes bellicosus (Brachyura: Portunidae) from the eastern Pacific Ocean. Mar Biol 146, 559–569
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49, 1280–1283
Reynolds J, Weir BS, Cockerham CC (1983) Estimation for the coancestry coefficient: basis for a short-term genetic distance. Genetics 105, 767–779
Roman J, Palumbi SR (2004) A global invader at home: population structure of the green crab, Carcinus maenas, in Europe. Mol Ecol 13, 2891–2898
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN: A Software for Population Genetics Data Analysis. Version 2.000 (Geneva, Switzerland: University of Geneva)
Schubart CD, Cuesta JA, Rodrııguez A (2001) Molecular. phylogeny of the crab genus Brachynotus (Brachyura: Varunidae) based on the 16S rRNA gene. Hydrobiologia 449, 41–46
Seeb JE, Kruse GH, Seeb LW, Weck RG (1989) Genetic structure of red king crab stocks in Alaska facilitates enforcement of fishing regulations. In: Proceedings of the International Symposium on King and Tanner Crabs, Anchorage, Alaska, November 28–30, 1989. Alaska Sea Grant College Program, Fairbanks, pp 491–502
Seeb LW, Grant S, Templin B, Kretschmer E, Olsen J (2001) Crab genetics. In: King and Tanner Crab Research in Alaska; Final Report. Alaska Department of Fish and Game. Division of Commercial Fisheries, pp 14–28
Senju T (1999) The Japan Sea intermediate water; its characteristics and circulation. J Oceanogr 55, 111–122
Sugaya T, Ikeda M, Taniguchi N (2002) Relatedness structure estimated by microsatellites DNA and mitochondrial DNA polymerase chain reaction—restriction fragment length polymorphisms analyses in the wild population of kuruma prawn Penaeus japonicus. Fish Sci 68, 793–802
Takayanagi S, Utoh H, Yorita K, Ueda Y (1999) Distribution and abundance of larvae of the atelecyclid crabs, Erimacrus isenbeckii (BRANDT) and Telmessus cheiragonus (TILESIUS) in Funka Bay and adjacent waters during April to early June 1989–1992. Sci Rep Hokkaido Fish Exp Stn 55, 79–87 (in Japanese with English abstract)
Tang BP, Zhou KY, Song DX, Yang G, Dai AY (2003) Molecular systematics of the Asian mitten crabs, genus Eriocheir (Crustacea: Brachyura). Mol Phylogenet Evol 29, 309–316
Uthicke S, Benzie JAH (2003) Gene flow and population. history in high dispersal marine invertebrates: mtDNA. analysis of Holothuria nobilis (Echinodermata: Holothuroidea) populations from the Indo-Pacific. Mol Ecol 12, 2635–2648
Watanabe K (1964) Hydrographic and meteorological conditions around Hokkaido. Bull Coastal Oceanogr 3, 23–30 (in Japanese)
Waters JM, Roy MS (2004) Phylogeography of a high-dispersal New Zealand sea-star: does upwelling block gene-flow? Mol Ecol 13, 2797–2806
Yamasaki I, Yoshizaki G, Yokota M, Strüssmann CA, Watanabe S (2006) Mitochondrial DNA variation and population structure of the Japanese mitten crab Eriocheir japonica. Fish Sci 72, 299–309
Yanagimoto T (2004) Geographic population subdivision of the Japanese sandfish, Arctoscopus japonicus, inferred from PCR-RFLP analysis on mtDNA. Nippon Suisan Gakkaishi 70, 583–591 (in Japanese with English abstract)
Acknowledgments
We thank Tomoaki Goto, Iwate Fisheries Technology Center, and the research ship crew of Iwate-maru and Kitakami-maru at the same institution for providing samples from the Pacific Ocean off the Iwate Prefecture, Honshu. This study was supported partly by Grant-in-Aid for 21st Century COE Program from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and by the Northern Advancement Center for Science and Technology, Hokkaido, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Azuma, N., Kunihiro, Y., Sasaki, J. et al. Genetic Variation and Population Structure of Hair Crab (Erimacrus isenbeckii ) in Japan Inferred from Mitochondrial DNA Sequence Analysis. Mar Biotechnol 10, 39–48 (2008). https://doi.org/10.1007/s10126-007-9033-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-007-9033-1