Abstract
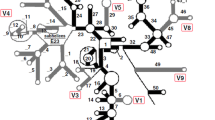
It is at present difficult to accurately position gaps in sequence alignment and to determine substructural homology in structure alignment when reconstructing phylogenies based on highly divergent sequences. Therefore, we have developed a new strategy for inferring phylogenies based on highly divergent sequences. In this new strategy, the whole secondary structure presented as a string in bracket notation is used as phylogenetic characters to infer phylogenetic relationships. It is no longer necessary to decompose the secondary structure into homologous substructural components. In this study, reliable phylogenetic relationships of eight species in Pectinidae were inferred from the structure alignment, but not from sequence alignment, even with the aid of structural information. The results suggest that this new strategy should be useful for inferring phylogenetic relationships based on highly divergent sequences. Moreover, the structural evolution of ITS1 in Pectinidae was also investigated. The whole ITS1 structure could be divided into four structural domains. Compensatory changes were found in all four structural domains. Structural motifs in these domains were identified further. These motifs, especially those in D2 and D3, may have important functions in the maturation of rRNAs.




Similar content being viewed by others
References
Bafna V, Muthukrishnan S, Ravi R (1995) “Computing similarity between RNA strings.” In: Proceedings of the 6th Symposium on Combinatorial Pattern Matching, Lecture Notes in Computer Science 937, Galil Z, Ukkonen E, eds. (Berlin: Springer-Verlag), pp 1–16
M Barucca E Olmo S Schiaparelli A Canapa (2004) ArticleTitleMolecular phylogeny of the family Pectinidae (Mollusca: Bivalvia) based on mitochondrial 16S and 12S rRNA genes Mol Phylogenet Evol 31 89–95 Occurrence Handle10.1016/j.ympev.2003.07.003
B Billoud M Guerrucci M Masselot JS Deutsch (2000) ArticleTitleCirripede phylogeny using a novel approach: molecular morphometrics Mol Biol Evol 17 1435–1445
G Caetano-Anolles (2002) ArticleTitleEvolved RNA secondary structure and the rooting of the universal tree of life J Mol Evol 54 333–345
CA Chen C Chang NV Wei C Chen Y Lein H Lin C Dai CC Wallace (2004) ArticleTitleSecondary structure and phylogenetic utility of the ribosomal internal transcribed spacer 2 (ITS2) in scleractinian corals Zool Stud 43 759–771
AW Coleman RM Preparata B Mehrotra JC Mai (1998) ArticleTitleDerivation of the secondary structure of the ITS-1 transcript in Volvocales and its taxonomical correlations Protist 149 135–146 Occurrence Handle10.1016/S1434-4610(98)70018-5
R Durbin S Eddy A Krogh G Mitchison (1998) Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids Cambridge University Press Cambridge, UK 260–298
M Fan BP Currie RR Gutell MA Ragan A Casadevall (1994) ArticleTitleThe 16S-like, 5.8S and 23S-like rRNAs of the two varieties of Cryptococcus neoformans: sequence, secondary structure, phylogenetic analysis and restriction fragment polymorphisms J Med Vet Mycol 32 163–180
W Fontana DAM Konings PF Stadler P Schuster (1993) ArticleTitleStatistics of RNA secondary structures Biopolymers 33 1389–1404 Occurrence Handle10.1002/bip.360330909
LR Goertzen JJ Cannone RR Gutell RK Jansen (2003) ArticleTitleITS secondary structure derived from comparative analysis: implications for sequence alignment and phylogeny of the Asteraceae Mol Phylogenet Evol 29 216–234 Occurrence Handle10.1016/S1055-7903(03)00094-0
AA Gontcharov M Melkonian (2005) ArticleTitleMolecular phylogeny of Staurastrum Meyen ex Ralfs and related genera (Zygnematophyceae, Streptophyta) based on coding and noncoding rDNA sequence comparisons J Phycol 41 887–899 Occurrence Handle10.1111/j.0022-3646.2005.04165.x
J Gorodkin LJ Heyer S Brunak GD Stormo (1997) ArticleTitleDisplaying the information contents of structural RNA alignments: the structure logos Comput Appl Biosci 13 583–586
M Gottschling J Plotner (2004) ArticleTitleSecondary structure models of the nuclear internal transcribed spacer regions and 5.8S rRNA in Calciodinelloideae (Peridiniaceae) and other dinoflagellates Nucl Acids Res 32 307–315 Occurrence Handle10.1093/nar/gkh168
M Gottschling HH Hilger M Wolf N Diane (2001) ArticleTitleSecondary structure of the ITS1 transcript and its application in a reconstruction of the phylogeny of Boraginales Plant Biol 3 629–636 Occurrence Handle10.1055/s-2001-19371
MA Hershkovitz LA Lewis (1996) ArticleTitleDeep-level diagnostic value of the rDNA-ITS region Mol Biol Evol 13 1276–1295
RE Hickson C Simon A Cooper GS Spicer J Sullivan D Penny (1996) ArticleTitleConserved sequence motifs, alignment, and secondary structure for the third domain of animal 12S rRNA Mol Biol Evol 13 150–169
DM Hillis MT Dixon (1991) ArticleTitleRibosomal DNA: molecular evolution and phylogenetic inference Q Rev Biol 66 411–453 Occurrence Handle10.1086/417338
IL Hofacker (2003) ArticleTitleVienna RNA secondary structure server Nucl Acids Res 31 3429–3431 Occurrence Handle10.1093/nar/gkg599
IL Hofacker W Fontana PF Stadler S Bonhoeffer M Tacker P Schuster (1994) ArticleTitleFast folding and comparison of RNA secondary structures Monatsh Chem 125 167–188 Occurrence Handle10.1007/BF00818163
IL Hofacker M Fekete PF Stadler (2002) ArticleTitleSecondary structure prediction for aligned RNA sequences J Mol Biol 319 1059–1066 Occurrence Handle10.1016/S0022-2836(02)00308-X
SR Holbrook (2005) ArticleTitleRNA structure: the long and the short of it Curr Opin Struc Biol 15 302–308 Occurrence Handle10.1016/j.sbi.2005.04.005
GC Hung NB Chilton I Beveridge RB Gasser (1999) ArticleTitleSecondary structure model for the ITS-2 precursor rRNA of strongyloid nematodes of equids: implications for phylogenetic inference Int J Parasitol 29 1949–1964 Occurrence Handle10.1016/S0020-7519(99)00155-1
A Insua MJ Lopez-Pinon R Freire J Mendez (2003) ArticleTitleSequence analysis of the ribosomal DNA internal transcribed spacer region in some scallop species (Mollusca: Bivalvia: Pectinidae) Genome 46 595–604 Occurrence Handle10.1139/g03-045
T Jiang G Lin B Ma K Zhang (2002) ArticleTitleA general edit distance between two RNA structures J Comput Biol 9 371–388 Occurrence Handle10.1089/10665270252935511
M Kimura (1980) ArticleTitleA simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences J Mol Evol 16 111–120 Occurrence Handle10.1007/BF01731581
KM Kjer (1995) ArticleTitleUse of rRNA secondary structure in phylogenetic studies to identify homologous positions: an example of alignment and data presentation from the frogs Mol Phylogenet Evol 4 314–330 Occurrence Handle10.1006/mpev.1995.1028
S Kumar K Tamura M Nei (2004) ArticleTitleMEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment Brief Bioinform 5 150–163 Occurrence Handle10.1093/bib/5.2.150
C Leblanc B Kloareg S Loiseaux-Degoer C Boyen (1995) ArticleTitleDNA sequence, structure, and phylogenetic relationship of the mitochondrial small-subunit rRNA from the red alga Chondrus crispus (Gigartinales rhodophytes) J Mol Evol 41 196–202 Occurrence Handle10.1007/BF00170673
G Lin LC Lo ZY Zhu F Feng R Chou GH Yue (2006) ArticleTitleThe complete mitochondrial genome sequence and characterization of single-nucleotide polymorphisms in the control region of the Asian seabass (Lates calcarifer) Mar Biotechnol 8 71–79 Occurrence Handle10.1007/s10126-005-5051-z
JS Liu CL Schardl (1994) ArticleTitleA conserved sequence in internal transcribed spacer 1 of plant nuclear rRNA genes Plant Mol Biol 26 775–778 Occurrence Handle10.1007/BF00013763
RJ Machida MU Miya MM Yamauchi M Nishida S Nishida (2004) ArticleTitleOrganization of the mitochondrial genome of Antarctic krill Euphausia superba (Crustacea: Malacostraca) Mar Biotechnol 6 238–250 Occurrence Handle10.1007/s10126-003-0016-6
JC Mai AW Coleman (1997) ArticleTitleThe internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants J Mol Evol 44 258–271 Occurrence Handle10.1007/PL00006143
BT Maynard LJ Kerr JM McKiernan ES Jansen PJ Hanna (2005) ArticleTitleMitochondrial DNA sequence and gene organization in Australian backup abalone Haliotis rubra (leach) Mar Biotechnol 7 645–658 Occurrence Handle10.1007/s10126-005-0013-z
JS McCaskill (1990) ArticleTitleThe equilibrium partition function and base pair binding probabilities for RNA secondary structure Biopolymers 29 1105–1119 Occurrence Handle10.1002/bip.360290621
B Michot N Joseph S Sazan JP Bachellerie (1999) ArticleTitleEvolutionary conserved structural features in the ITS2 of mammalian pre-rRNAs and potential interactions with the snoRNA U8 detected by comparative analysis of new mouse sequences Nucl Acids Res 27 2271–2282 Occurrence Handle10.1093/nar/27.11.2271
CA Milbury PM Gaffney (2005) ArticleTitleComplete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica Mar Biotechnol 7 697–712 Occurrence Handle10.1007/s10126-005-0004-0
AD Miller NP Murphy CP Burridge CM Austin (2005) ArticleTitleComplete mitochondrial DNA sequences of the decapod crustaceans Pseudocarcinus gigas (Menippidae) and Macrobrachium rosenbergii (Palaemonidae) Mar Biotechnol 7 339–349 Occurrence Handle10.1007/s10126-004-4077-8
DA Morrison JT Ellis (1997) ArticleTitleEffects of nucleotide sequence alignment on phylogeny estimation: a case study of 18S rDNAs of Apicomplexa Mol Biol Evol 14 428–441
W Musters K Boon CAFM Sande Particlevan der H Heerikhuizen Particlevan RJ Planta (1990) ArticleTitleFunctional analysis of transcribed spacers of yeast ribosomal DNA EMBO J 9 3989–3996
GJ Olsen (1988) ArticleTitlePhylogenetic analysis using ribosomal RNA Methods Enzymol 164 793–812 Occurrence Handle10.1016/S0076-6879(88)64084-5
L Podsiadlowski T Bartolomaeus (2005) ArticleTitleOrganization of the mitochondrial genome of mantis shrimp Pseudosquilla ciliate (Crustacea: Stomatopoda) Mar Biotechnol 7 618–624 Occurrence Handle10.1007/s10126-005-0017-8
R Reedy LI Rothblum CS Subrahmanyam M Liu D Henning B Cassidy H Busch (1983) ArticleTitleThe nucleotide sequence of 8S RNA bound to preribosomal RNA of Novikoff hepatoma J Biol Chem 258 584–589
N Saitou M Nei (1987) ArticleTitleThe neighbor-joining method: a new method for reconstructing phylogenetic trees Mol Biol Evol 4 406–425
TD Schneider RM Stephens (1990) ArticleTitleSequence logos: a new way to display consensus sequences Nucl Acids Res 18 6097–6100 Occurrence Handle10.1093/nar/18.20.6097
BA Shapiro (1988) ArticleTitleAn algorithm for comparing multiple RNA secondary structures CABIOS 4 381–393
BA Shapiro K Zhang (1990) ArticleTitleComparing multiple RNA secondary structures using tree comparison CABIOS 6 309–318
BW Tague SA Gerbi (1984) ArticleTitleProcessing of the large rRNA precursor: two proposed categories of RNA-RNA interactions in eukaryotes J Mol Evol 20 362–367 Occurrence Handle10.1007/BF02104742
JD Thompson TJ Gibson F Plewniak F Jeanmougin DG Higgins (1997) ArticleTitleThe Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools Nucl Acids Res 24 4876–4882 Occurrence Handle10.1093/nar/25.24.4876
TA Titus DR Frost (1996) ArticleTitleMolecular homology assessment and phylogeny in the lizard family Opluridae (Squamata: Iguania) Mol Phylogenet Evol 6 49–62 Occurrence Handle10.1006/mpev.1996.0057
CAFM Sande Particlevan der M Kwa RW Nues Particlevan H Heerikhuizen Particlevan HA Raue RJ Planta (1992) ArticleTitleFunctional analysis of internal transcribed spacer 2 of Saccharomyces cerevisiae ribosomal DNA J Mol Biol 223 899–910 Occurrence Handle10.1016/0022-2836(92)90251-E
RW Nues Particlevan JMJ Rientjes CAFM Sande Particlevan der SF Zerp C Sluiter J Venema RJ Planta AHA Raue (1994) ArticleTitleSeparate structural elements within internal transcribed spacer 1 of Saccharomyces cerevisiae precursor ribosomal RNA direct the formation of 17S and 26S rRNA Nucl Acids Res 22 912–919 Occurrence Handle10.1093/nar/22.6.912
RW Nues Particlevan JMJ Rientjes SA Morre E Mollee RJ Planta J Venema HA Raue (1995) ArticleTitleEvolutionarily conserved structural elements are critical for processing of internal transcribed spacer 2 from Saccharomyces cerevisiae precursor ribosomal RNA J Mol Biol 250 24–36 Occurrence Handle10.1006/jmbi.1995.0355
JHG Schulenburg Particlevonder U Englisch JW Wagele (1999) ArticleTitleEvolution of ITS1 rDNA in the Digenea (Platyhelminthes: Trematoda): 3′ end sequence conservation and its phylogenetic utility J Mol Evol 48 2–12 Occurrence Handle10.1007/PL00006441
JHG Schulenburg Particlevonder JM Hancock A Pagnamenta JJ Sloggett MEN Majerus GDD Hurst (2001) ArticleTitleExtreme length and length variation in the first ribosomal internal transcribed spacer of ladybird beetles (Coleoptera: Coccinellidae) Mol Biol Evol 18 648–660
Wang Z, Zhang K (2001) “Alignment between two RNA structures.” In: Proceedings of the 26th Symposium on the Mathematical Foundations of Computer Science, Lecture Notes in Computer Science 2136, Sgall J, Pultr A, Kolman P, eds. (London: Springer-Verlag), pp 690–702
MM Yamauchi MU Miya RJ Machida M Nishida (2004) ArticleTitlePCR-based approach for sequencing mitochondrial genomes of decapod crustaceans, with a practical example from kuruma prawn (Marsupenaeus japonicus) Mar Biotechnol 6 419–429 Occurrence Handle10.1007/s10126-003-0036-2
Zhang K (1998) “Computing similarity between RNA secondary structures.” In: IEEE International Joint Symposium on Intelligence and Systems, Liu Z, ed. (Los Alamitos, CA: IEEE Computer Society Press), pp 126–132
Zhang K, Wang L, Ma B (1999) “Computing similarity between RNA structures.” In: Proceedings of the 10th Symposium on Combinatorial Pattern Matching, Lecture Notes in Computer Science 1645, Crochemore M, Paterson M, eds. (London: Springer-Verlag), pp 281–293
Acknowledgments
The authors thank Prof. Guanpin Yang for reviewing the manuscript. This work was supported by grants from Hi-Tech Research and Development Program of China (2005AA603220), National Natural Science Foundation of China (30300268) and the Key Laboratory of Mariculture of Ministry of Education, Ocean University of China (200417).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, S., Bao, Z., Li, N. et al. Analysis of the Secondary Structure of ITS1 in Pectinidae: Implications for Phylogenetic Reconstruction and Structural Evolution. Mar Biotechnol 9, 231–242 (2007). https://doi.org/10.1007/s10126-006-6113-6
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-006-6113-6