Abstract
To identify lobster phyllosoma larvae of the genus Panulirus occurring in waters adjacent to Japan, genetic variation within and between 10 Indo-Pacific lobster species was investigated using restriction fragment length polymorphism (RFLP) analysis for the 1300-base pair mitochondrial cytochrome oxidase I (COI) gene. RFLP analysis using two endonucleases (AluI and TaqI) enabled discrimination of all species, including the P. longipes complex. The diagnostic DNA markers, supplemented with nucleotide sequence analysis, were applied to 44 mid- to late-stage phyllosoma larvae (7.4 to 27.7 mm in body length) collected in the northwestern Pacific. These samples were unexpectedly variable in species composition, comprising P. japonicus (n = 16), P. longipes bispinosus (21), P. longipes longipes (1), P. “aka” (1), and P. penicillatus (5). Comparison of larval size at similar stages revealed that P. l. bispinosus larvae were significantly larger than P. japonicus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spiny lobsters are one of the world's most valuable fisheries resources. The genus Panulirus, comprising 19 or more species, is the largest group in the family Palinuridae (George and Main, 1967; Holthuis, 1991; McWilliam, 1995), and all species are highly prized in many countries. Much attention has been paid to biological investigation of the bottom-living phase of the young and adult stages, while biological studies of the planktonic stage are scarce. This is mainly due to taxonomic uncertainty at the larval stage and to small sample sizes of wild-caught larvae. Larvae of the spiny lobster, called phyllosoma, are known to be peculiar, having highly transparent and dorsoventrally compressed bodies and a very long planktonic period extending from several months to more than a year (Chittleborough and Thomas, 1969; Inoue, 1981; Kittaka and Kimura, 1989; Yamakawa et al., 1989; Matsuda and Yamakawa, 2000). This long larval period accompanied by morphological change has made species identification of phyllosoma larvae very difficult, especially where multiple species share similar distribution ranges.
The Japanese spiny lobster, Panulirus japonicus, is the most abundant lobster in the temperate coastal area of Japan, followed by Panulirus longipes, which is widely distributed in tropical to subtropical waters of the Indo-Pacific (Holthuis, 1991). These two species are closely related and belong to species-group I or “P. japonicus” group (George and Holthuis, 1965; George and Main, 1967). Johnson (1971) pointed out that the larvae of the P. japonicus group may be very similar or indistinguishable from one another in morphology. Following Gurney's description (1936), Oshima (1942) reported that two Forms (E and F) occur in Japanese waters. Later, Murano (1971) observed five Forms (A to E) of late-stage phyllosoma larvae of Panulirus in waters adjacent to Japan and suggested his Form A corresponded to Oshima's Form F. Murano (1971), and subsequently Nonaka et al. (1989) suggested that phyllosoma larvae of P. japonicus and P. longipes both belong to Form A, but exact species identification was not possible. Sekiguchi (1986) proposed that late stage phyllosoma larvae of P. japonicus and P. longipes can be discriminated by the ratio of cephalic shield and thorax widths. Subsequently, however, the utility of this ratio in identifying these two species was questioned by Inoue and Sekiguchi (2001). Observations from complete larval rearing indicated that subfinal and final stages of phyllosoma larvae of P. japonicus and P. longipes conformed almost exactly with Murano's Form A and that there was very little morphological difference between these two species throughout their larval stages (Inoue, 1981; Matsuda and Yamakawa, 2000).
Morphological and recent molecular analyses have revealed P. longipes to be a species complex, making morphological identification at the larval stage even more difficult. Holthuis (1991) described two subspecies, Panulirus longipes longipes with spotted-legs and P. longipes femoristriga with striped-legs, in P. longipes. Subsequent taxonomic studies have revealed P. l. femoristriga is also a species complex, in which one subspecies and two species have been recognized as P. l. bispinosus (=P. l. “shirahige”), P. femorstriga (=P. albifragellum) and P. “aka” (Sekiguchi, 1991; Chan and Chu, 1996; George, 1997; Chan and Ng, 2001). Molecular phylogenetic analyses using mitochondrial DNA sequences (Ptacek et al., 2001; Ravago and Juinio-Meñez, 2003) have indicated close but distinct relationships between P. l. longipes and P. l. bispinosus and thedistinct specific status of P. femoristriga, while P. “aka” has been not included in the molecular analyses. Although phyllosoma larvae of all of thesespecies may occur in Japanese waters, definitive morphological identification has been not possible even for the final stage (McWilliam, 1995; Yoshimura et al., 1999; Matsuda and Yamakawa, 2000; Inoue and Sekiguchi, 2001; Yoshimura et al., 2002). Yoshimura et al. (2002) suggested that morphological and DNA analyses for wild larvae, including those of the P. longipes complex, should bedeveloped for advancing the larval study of P.japonicus.
DNA analysis can support species identification, and in fact restriction fragment length polymorphism (RFLP) and/or direct nucleotide sequencing analyses based on polymerase chain reaction (PCR) methods have become conventional and practical for identifying fish and crustacean species at all stages of life cycle (Silberman and Walsh, 1992; Chow et al., 1993, 2003; Imai et al., 2004). Furthermore, PCR amplification can be performed for any DNA region from a tiny piece of tissue, thus preserving the sample for subsequent morphological investigation.
In this study, we present diagnostic DNA markers able to identify 10 spiny lobster species of the genus Panulirus occurring in the Indo-Pacific, and report the results of species identification for mid- to late-stage phyllosoma larvae collected in the northwestern Pacific.
Materials and Methods
Lobster Samples and DNA Extraction
Collection location and sample sizes of the standard adult specimens are shown in Table 1. Adult individuals purchased at local landing sites or caught by local fisherman were transferred to the laboratory alive orfrozen. Tissue samples collected by foreign organizations were transferred to the laboratory in ethanol. Forty-four phyllosoma larvae (designated L1 to L45; L7 was lacking) collected by the RV Yoko-Maru using an Isaacs-Kidd Midwater Trawl (IKMT) net from January 14 to 30, 2002 in the northwestern Pacific (27°30′–30°27′N: 133°20′–135°E) were fixed in ethanol on board and transferred to the laboratory. The phyllosoma larvae were morphologically assigned to Forms A (n = 39) and C (n = 5) according to Murano (1971), and all Form A larvae were staged according to Inoue (1981) and Matsuda and Yamakawa (2000). Using a biopsy instrument (Disposable Biopsy Punch, Kai Group Ltd., Japan), a small piece of tissue (2 mm diameter) was punched from the thorax region of the larvae and stored in ethanol until use.
Tissue samples from adult specimens were finely minced and those from the phyllosoma larvae were homogenized in 1.5 ml microcentrifuge tubes using a Teflon pestle. Crude DNA was extracted using a DNA extraction kit (GenomicPrep Cells and Tissue DNA Isolation Kit, Amersham Bioscience).
PCR Amplification
Silberman and Walsh (1992) demonstrated successful discrimination among phyllosoma larvae of three Panulirus species in the western Atlantic using restriction analysis on 28S rDNA amplified by PCR. However, we failed to discriminate two closely related species (P. l. bispinosus and P. l. longipes) using 11 endonucleases (AflIII, DdeI, HhaI, HinfI, MseI, MspI, NlaIII, NlaIV, RsaI, Sau96I, and TaqI) on this gene. Therefore, mitochondrial cytochrome oxidase I gene (COI) was adopted for RFLP analysis, since partial COI sequences of almost all Panulirus species have been reported and are available from the GenBank database (Ptacek et al., 2001; Ravago and Juinio-Meñez, 2003). Primers for PCR amplification are shown in Table 2. All forward primers were designed to anneal to an identical site near the 5′-end, and reverse primers were designed to anneal at the 3′ region of the COI. First, a single pair of primers (COI65F1 and COI1342R1) was applied to all samples, and subsequently all eight primers were mixed and used for samples that were not well amplified by the first pair of primers. We observed that LA Taq polymerase with GC buffer (TAKARA Ltd., Kyoto) greatly increased amplification efficiency for the spiny lobster COI compared to the standard Taq DNA polymerase. PCR amplification for all specimens was carried out in a 10 μl reaction mixture containing 5 μl of GC buffer, 1 mM of each dNTP, primers (1 μM each for single pair and 0.2μM each for a mixed one), 0.5 unit of LA Taq polymerase, and DNA template. The reaction mixtures were preheated at 95°C for 3 min followed by 30 cycles of amplification (at 95°C for 1 min, 55°C for 30 s, and 72°C for 1.5 min) with a final extension at 72°C for 5 min. The amplification was usually very strong, and 10 to 20 μl of sterilized water was added to dilute the PCR products prior to subsequent analysis.
RFLP and Nucleotide Sequence Analyses
We searched for restriction site differences among lobster species using published nucleotide sequence data reported by Ptacek et al. (2001) and Ravago and Juinio-Meñez (2003), and selected two endonucleases (AluI and TaqI) that provided diagnostic species patterns. The PCR products were directly digested by these enzymes and electrophoresed on a 2.5% agarose gel (Biogel, BIO101, Inc.) for 2 to 3 h, followed by ethidium bromide staining and photographic recording.
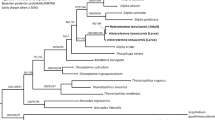
Nucleotide sequence analysis was performed on phyllosoma samples showing different RFLP profiles from those of the adult standards. In addition, two adult P. “aka” individuals were analyzed, since the sequence of this species has not been reported. Oligonucleotide primers were removed from the PCR products using ExoSAP-IT (Amersham Biosciences) to prepare a DNA template. Sequences were generated on an automated sequencer (ABI Prism310) using the ABI Big-dye Ready Reaction kit following the standard cycle sequencing protocol. Since previous studies on lobster phylogeny adopted Kimura's two-parameter distance (K2P)(Ptacek et al., 2001; Ravago and Juinio-Meñez, 2003), we incorporated these published sequence data to calculate K2P distance and constructed neighbor-joining (NJ) tree (Saitou and Nei, 1987) using MEGA ver. 3 (Kumar et al., 2004). The max-mini branch-and-bound search algorithm implemented in MEGA was adopted for parsimony analysis.
Results
RFLP within and between Standard Species Samples
The size of amplified fragments was estimated to be approximately 1300 base pairs (bp), and no apparent size variation was observed among 107 adult standard specimens. RFLP within and between species are shown in Figure 1, in which AluI and TaqI digestions detected 19 and 14 restriction types, respectively. Size and distribution of fragments in each restriction type observed in AluI and TaqI digestions can be obtained from http://www.enyo.affrc.go.jp/chow/LobsterRFLP.htm. Composite haplotypes are summarized in Table 3. Intraspecific variation was observed in all species except P. “aka” and P. versicolor. Almost all species showed distinct restriction profiles from each other except for P. l. bispinosus and P. l. longipes, which shared an identical profile in AluI digestion (type A5), and P. “aka” and P. marginatus which shared an identical profile in TaqI digestion (type T8) (see Figure 1). No composite haplotype was observed to be shared by different species (Table3), indicating that all standard species of the genus Panulirus used in this study could be readily discriminated using these two restriction enzymes.
AluI (top) and TaqI (bottom) restriction profiles of the mitochondrial cytochrome oxidase I (COI) gene fragment of standard lobster specimens. The sizes of the molecular markers (M: 100 bp DNA ladder, New England Biolab) are shown in the left margin. See Table 1 for abbreviations of spiny lobster species. U, Undigested PCR product.
Identification of Phyllosoma Larvae
The restriction analysis indicated that the morphological assignments were roughly correct. In addition, the restriction analysis further delineated species identity, and found several instances in which larvae showed novel restriction profiles not observed in the adult standards. Among 39 Form A larvae, species identification using RFLP analysis was accomplished for 34 larvae, comprising P. japonicus (n = 14), P. l. bispinosus (18), P. l. longipes (1), and P. “aka” (1). Of the remaining five Form A larvae, two (L16 and 32) had novel restriction profiles in AluI digestion (designated as type A20) but shared an identical profile with the P. japonicus standard in TaqI digestion (type T1). The other three Form A larvae (L2, 22, and 31) shared identical restriction profiles with P. l. bispinosus and P. l. longipes in AluI digestion (type A5) but had novel profiles in TaqI digestion (types T15 and 16). In five Form C larvae, four individuals shared identical restriction profiles with P. penicillatus standards. One Form C larva (L6) had a novel profile in AluI digestion (type A21) but shared an identical profile with P. penicillatus in TaqI digestion (type T9). Nucleotide sequences of five of the six larvae (L2, 6, 16, 22, and 32), L21 with the P. l. longipes restriction profile and L45 with the P. “aka” restriction profile, were determined and compared with published data. Nucleotide sequence of one larva (L31) sharing the identical restriction profiles with L22 was not analyzed. An entire COI sequence of P. japonicus was obtained from whole mtDNA sequence data reported by Yamauchi et al. (2002). Ptacek et al. (2001) sequenced near the 5′ upstream region of COI (approximately 640 bp) for almost all the Panulirus species. Ravago and Juinio-Meñez (2003) analyzed a further downstream region (approximately 560 bp) of fewer species but focused intensively on the P. longipes complex. Sarver et al. (1998) reported P. cygnus and P. marginatus COI sequences corresponding to the region examined by Ravago and Juinio-Meñez (2003). Nucleotide sequences of Jasus edwardsii and Parribacus antarcticus were derived from Ptacek et al. (2001) and Ravago and Juinio-Meñez (2003), respectively, as out group species. Homology investigation and alignment among these published sequences and our data allowed us to sample 483 bp in the upstream region and 338 bp in the downstream region of COI for phylogenetic analysis. Nucleotide sequences of these larvae and two P. “aka” standards are available in DDBJ under accession numbers AB193071 to AB193088. Neighbor-joining (NJ) and maximum parsimony (MP) trees constructed using upstream and downstream region sequence data are shown in Figure 2. All trees constructed using upstream and downstream region data supported two major lineages (groups I plus II and III plus IV as defined by George and Main, 1967). Although relationships among species within each lineage varied as observed byPtacek et al. (2001) and Ravago and Juinio-Meñez (2003), the close (mean K2P = 0.051 ± 0.01 and 0.063 ± 0.013 for upstream and downstream regions, respectively) but distinct relationships between P. l. bispinosus and P. l. longipes were evident. Mean nucleotide sequence divergences (K2P) between P. “aka” and other species were 0.230 ± 0.025 (SE) for the upstream and 0.198 ± 0.02 for the downstream sequences, comparable or even larger than those between other species. The deep branch of the P. “aka” cluster and the large sequence divergence from the other species may well support the species status. Assignment of all six larvae unidentified by RFLP assay was unambiguously established in all tree construction methods. Thus, restriction analysis supplemented by the nucleotide sequencing indicated our phyllosoma sample to consist of 16 P. japonicus, 21 P. l. bispinosus, one P.l. longipes, one P. “aka,” and 5 P. penicillatus.
Neighbor-joining (NJ) (A, C) and maximum parsimony (MP) (B, D) phylogenetic trees drawn using the upstream (A, B) and downstream (C, D) region sequence data of COI. Assignments of all larvae (undetermined by RFLP analysis) were established, and the species status of P. “aka” is strongly supported. Published sequence data are derived from \(^{\Psi } \)Sarver et al. (1998), *Ptacek et al. (2001), $Yamauchi et al. (2002), and \(^{\phi } \)Ravago and Juinio-Meñez (2003). Bootstrapping was run with 1000 replicates and values over 50% are shown at the nodes.
Heterogeneous larval body size between larvae of Form A species was observed. Based on the size and morphology, the larval stage of P. japonicus was determined to be VI to VII, comprising 10 and 6 individuals, respectively. Body length of stage VII P. japonicus larvae ranged from 12.6 to 15.5 mm with a mean of 13.8 mm ± 1.03 (SD). Phyllosoma larvae of P. l. bispinosus consisted of 5 VII, 15 VIII, and 1 IX stage larvae, in which body length of stage VII larvae ranged from 15.1 to 20.0 mm with a mean of 17.2 mm ± 2.29, significantly larger than that of P.japonicus (Mann-Whitney's U test, P < 0.05). Stage VIII larvae of 15 P. l. bispinosus ranged from18.6 to 27.3 mm with a mean of 23.1 mm ± 2.19, and that of stage IX larva was 27.6 mm. Body length of one stage IX larva of P. l. longipes was 27.7 mm, and that of one stage VII larva of P. “aka” was 15.0 mm.
Discussion
The present study has introduced a simple RFLP assay to identify Pacific spiny lobster species of the genus Panulirus. Investigation of intraspecific variation may be essential for species identification, and larger sample sizes for several species would be useful to further substantiate the RFLP markers. However, the probability of mis-identification based on the RFLP analysis is very low, since substantial nucleotide sequence divergence has been observed among species of the genus Panulirus (Ptacek et al., 2001; Ravago and Juinio-Meñez, 2003; in this study) except for the two closely related subspecies (P. l. bispinosus and P. l. longipes). Relatively low levels of restriction site polymorphism within species and very little restriction profile sharing between species observed in the present study further corroborate the diagnostic utility of the RFLP markers presented here. Furthermore, the wide range of geographic locations represented in the standard samples in some species (see Table 1) may compensate for the limited sample size.
The present study is a step toward resolving problems in species identification for phyllosoma larvae of Panulirus lobsters and provides a breakthrough for studying distribution and transport of phyllosoma larvae. Inoue and Sekiguchi (2001) concluded all Form A phyllosoma larvae collected east of the Ryukyu Archipelago were P. japonicus. Despite the lack of morphological evidence for species identification, they also speculated on the transport and recruitment processes of phyllosoma larvae in this species. Phyllosoma larvae used in the present study were caught further to the north, but the species composition was unexpectedly variable in contrast to the implications of Inoue and Sekiguchi (2001). Interestingly, Form A individuals examined by Inoue and Sekiguchi (2001) were collected between March to July and consisted of two size groups (a subfinal stage ranging from 20 to 27 mm, and a final stage ranging from 29 to 34 mm), possibly corresponding to our identified larvae of P. japonicus (11.3 to 15.5 mm) and P. l. bispinosus (15.1 to 27.6 mm) collected earlier in the life cycle (i.e., January). In the present study, larval stages of P. japonicus and P. l. bispinosus were estimated to be stages VI and VII and VII to IX, respectively, and P. l. bispinosus larvae were significantly larger than P. japonicus. This is consistent with the observation in laboratory-reared phyllosoma larvae, in which larvae of P. longipes were found to be larger than those of P. japonicus at all stages (Inoue, 1981; Matsuda and Yamakawa, 2000). In contrast, larvae of P. longipes reared by Saisho and Nakahara (1960) were even smaller than those of P. japonicus. Staging of palinurid and scyllarid larvae may be arbitrary especially at later developmental stages (Johnson, 1971; Matsuda and Yamakawa, 2000), and the captive environment may significantly affect the growth of phyllosoma larvae (Matsuda and Yamakawa, 2000; Duggan and McKinnon, 2003). Nevertheless, size differences at similar larval stage observed in the present study may be key to primary sorting of Form A larvae occurring naturally in Japanese waters. Since our sample was collected within a limited area and season, more samples from a wider geographic range and a longer time span will be necessary to further investigate the dynamics of species composition and distribution of phyllosoma larvae.
References
TY Chan KH Chu (1996) ArticleTitleOn the different forms of Panulirus longipes femoristriga (Von Martens, 1872) (Crustacea: Decapoda: Palinuridae), with description of a new species J Nat Hist 30 367–387
TY Chan PKL Ng (2001) ArticleTitleOn the nomenclature of the commercially important spiny lobsters Panulirus longipes femoristriga (Von Martens, 1872), P. bispinosus Borradaile, 1899, and P. albiflagellum Chan and Chu, 1996 (Decapoda, Palinuridae) Crustaceana 74 123–127 Occurrence Handle10.1163/156854001505307
RG Chittleborough LR Thomas (1969) ArticleTitleLarval ecology of Western Australian crayfish, with notes upon other Panulirus larvae from the eastern Indian Ocean Austral J Mar Freshwat Res 20 199–223 Occurrence Handle10.1071/MF9690199
S Chow ME Clarke PJ Walsh (1993) ArticleTitlePCR-RFLP analysis on thirteen western Atlantic snappers (subfamily Lutjaninae): a simple method for species and stock identification Fish Bull 91 619–627
S Chow K Nohara T Tanabe T Itoh S Tsuji Y Nishikawa S Ueyanagi K Uchikawa (2003) ArticleTitleGenetic and morphological identification of larval and small juvenile tunas (Pisces: Scombridae) caught by a mid-water trawl in the western Pacific Bull Fish Res Agency 8 1–14
S Duggan AD McKinnon (2003) ArticleTitleThe early larval developmental stages of the spiny lobster Panulirus ornatus (Fabricius, 1798) cultured under laboratory conditions Crustaceana 76 313–332 Occurrence Handle10.1163/156854003765911711
RW George (1997) ArticleTitleTectonic plate movements and the evolution of Jasus and Panulirus spiny lobsters (Palinuridae) Mar Freshwat Res 48 1121–1130 Occurrence Handle10.1071/MF97202
RW George LB Holthuis (1965) ArticleTitleA revision of the Indo-West Pacific spiny lobsters of the Panulirus japonicus group Zool Verhand Leiden 72 1–36
RW George AR Main (1967) ArticleTitleThe evolution of spiny lobsters (Palinuridae): a study of evolution in the marine environment Evolution 21 803–820 Occurrence Handle10.2307/2406775
R Gurney (1936) ArticleTitleLarvae of Decapod Crustacea, Part III. Phyllosoma Discov Rep 12 400–440
LB Holthuis (1991) Marine Lobsters of the World: An Annotated and Illustrated Catalogue of Species of Interest to Fisheries Known to Date: FAO species Catalogue,Vol. 13 FAO Rome
H Imai J-H Cheng K Hamasaki K Numachi (2004) ArticleTitleIdentification of four mud crab species (genus Scylla) using ITS-1 and 16S rDNA markers Aquat Living Resour 17 31–34 Occurrence Handle10.1051/alr:2004007
M Inoue (1981) ArticleTitleStudies on the cultured phyllosoma larvae of the Japanese spiny lobster, Panulirus japonicus (V. Siebold) Special Rep Kanagawa Pref Fish Exp St 1 1–91
N Inoue H Sekiguchi (2001) ArticleTitleDistribution of late-stage phyllosoma larvae of Panulirus japonicus in the Kuroshio Subgyre Mar Freshwat Res 52 1201–1209 Occurrence Handle10.1071/MF01184
MW Johnson (1971) ArticleTitleOn palinurid and scyllarid lobster larvae and their distribution in the South China Sea (Decapoda, Palinuridea) Crustaceana 21 247–282 Occurrence Handle10.1163/156854071X00599
J Kittaka K Kimura (1989) ArticleTitleCulture of the Japanese spiny lobster Panulirus japonicus from egg to juvenile stage Nippon Suisan Gakkaishi 55 963–970
S Kumar K Tamura M Nei (2004) ArticleTitleMEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment Brief Bioinform 5 150–163 Occurrence Handle10.1093/bib/5.2.150
H Matsuda T Yamakawa (2000) ArticleTitleThe complete development and morphological changes of larval Panulirus longipes (Decapoda, Palinuridae) under laboratory conditions Fish Sci 66 278–293 Occurrence Handle10.1046/j.1444-2906.2000.00045.x
PS McWilliam (1995) ArticleTitleEvolution in the phyllosoma and puerulus phages of the spiny lobster genus Panulirus White J Crust Biol 15 542–557 Occurrence Handle10.2307/1548775
M Murano (1971) ArticleTitleFive forms of palinurid phyllosoma larvae from Japan Publ Seto Mar Biol Lab 19 17–25
M Nonaka M Hataya M Aoyama K Yamamoto (1989) ArticleTitleThe relative growth of Panulirus phyllosoma larvae in Japanese waters Nippon Suisan Gakkaishi 55 605–612
Y Oshima (1942) ArticleTitleOn the phyllosoma of the genus Panulirus Suisan Gakkaiho 9 36–44
MB Ptacek SK Sarver MJ Childress WF Herrnkind (2001) ArticleTitleMolecular phylogeny of the spiny lobster genus Panulirus (Decapoda: Palinuridae) Mar Freshwat Res 52 1037–1047 Occurrence Handle10.1071/MF01070
RG Ravago MA Juinio-Meñez (2003) ArticleTitlePhylogenetic position of the striped-legged forms of Panulirus longipes (A. Milne-Edwards, 1868) (Decapoda, Palinuridae) inferred from mitochondrial DNA sequences Crustaceana 75 1047–1059 Occurrence Handle10.1163/156854002763270455
T Saisho K Nakahara (1960) ArticleTitleOn the early development of phyllosomas of Ibacus ciliatus (von Siebold) and Panulirus longipes (A. Milne Edwards) Mem Fac Fish Kagoshima Univ 9 84–90
N Saitou M Nei (1987) ArticleTitleThe neighbour-joining method: a new method for reconstructing phylogenetic trees Mol Biol Evol 4 406–425
S Sarver JD Silberman PJ Walsh (1998) ArticleTitleMitochondrial DNA sequence evidence supporting the recognition of two subspecies or species of the Florida spiny lobster Panulirus argus J Crust Biol 18 177–186 Occurrence Handle10.2307/1549532
H Sekiguchi (1986) ArticleTitleIdentification of late-stage phyllosoma larvae of the scyllarid and palinurid lobsters in the Japanese waters Bull Jap Soc Sci Fish 52 1289–1294
H Sekiguchi (1991) ArticleTitleTwo forms of Panulirus longipes femoristriga (Crustacea, Palinuridae) from Ogasawara waters, Japan Proc Jpn Soc Syst Zool 44 15–25
JD Silberman PJ Walsh (1992) ArticleTitleSpecies identification of spiny lobster phyllosome larvae via ribosomal DNA analysis Mol Mar Biol Biotech 1 195–205
T Yamakawa M Nishimura H Matsuda A Tsujigado N Kamiya (1989) ArticleTitleComplete larval rearing of the Japanese spiny lobster Panulirus japonicus Nippon Suisan Gakkaishi 55 745
M Yamauchi M Miya M Nishida (2002) ArticleTitleComplete mitochondrial DNA sequence of the Japanese spiny lobster, P. japonicus Gene 295 89–96 Occurrence Handle10.1016/S0378-1119(02)00824-7
T Yoshimura H Yamakawa E Kozasa (1999) ArticleTitleDistribution of final stage phyllosoma larvae and free-swimming pueruli of Panulirus japonicus around the Kuroshio Current off southern Kyushu, Japan Mar Biol 133 293–306 Occurrence Handle10.1007/s002270050468
T Yoshimura K Morinaga S Shirai H Yamakawa (2002) ArticleTitlePalinurid phyllosoma larvae and their distribution in winter off the Pacific coast of Japan Fish Sci 68 194–197
Acknowledgments
We thank M. Childress and M.B. Ptacek and their colleague of the Lobster Phylogeny Project at Clemson University, R.G. Ravago and A. Juinio-Meñez of the Marine Science Institute, the University of Philippines, G. N. Permana and A. Nakazawa of the Gondol Research Institute of Mariculture, J.B. Wexler of the Inter-American Tropical Tuna Commission, and J. Uchiyama of the National Marine Fisheries Service, for generously providing invaluable lobster tissue samples and for their efforts to collect lobster samples. Some samples of adult lobster were kindly made available by Mr. K. Nishikiori and Mr. T. Yamamoto of the Tokyo Metropolitan Fisheries Experiment Station and Mr. T. Maekawa of the Maekawa Fishery Co., Ltd. We also like to thank the members of RV Yoko-Maru for assistance in sample collection, S. Clarke for reading and considerably improving this manuscript, and H. Hasegawa and M. Michibayashi, for their superb technical assistance in DNA analyses. This work was supported in part by grants from the Japanese Society for the Promotion of Science, the Ministry of Agriculture, Forestry, and Fisheries of Japan, and a Grant-in-Aid for Scientific Research on Priority Areas (B) from the Ministry of Education, Science, Sports and Culture.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Chow, S., Suzuki, N., Imai, H. et al. Molecular Species Identification of Spiny Lobster Phyllosoma Larvae of the Genus Panulirus from the Northwestern Pacific. Mar Biotechnol 8, 260–267 (2006). https://doi.org/10.1007/s10126-005-5151-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-005-5151-9