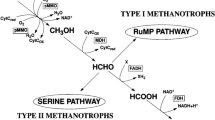
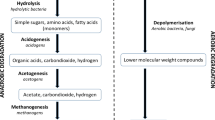
Abstract
Wetlands are the main natural sources of methane emissions, which make up a significant portion of greenhouse gas emissions. Such wetland patches serve as rich habitats for aerobic methanotrophs. Limited knowledge of methanotrophs from tropical wetlands widens the scope of study from these habitats. In the present study, a freshwater wetland in a tropical region in India was sampled and serially diluted to obtain methanotrophs in culture. This was followed by the isolation of methanotrophs on agarose-containing plates, incubated under methane: air atmosphere. Methanotrophs are difficult to cultivate, and very few cultures of methanotrophs are available from tropical wetlands. Our current study reports the cultivation of a diverse community of methanotrophs from six genera, namely, Methylomonas, Methylococcus, Methylomagnum, Methylocucumis (type I methanotrophs) along with Methylocystis, Methylosinus (type II methanotrophs). A high abundance of methanotrophs (106–1010 methanotrophs/g fresh weight) was observed in the samples. A Methylococcus strain could represent a putative novel species that was also isolated. Cultures of Methylomagnum and Methylocucumis, two newly described type I methanotrophs exclusively found in rice fields, were obtained. A large number of Methylomonas koyamae strains were cultured. Our study is pioneering in the documentation of culturable methanotrophs from a typical tropical wetland patch. The isolated methanotrophs can act as models for studying methanotroph-based methane mitigation from wetland habitats and can be used for various mitigation and valorization applications.



Similar content being viewed by others
References
Awala SI, Bellosillo LA, Gwak J-H, Nguyen N-L, Kim S-J, Lee B-H, Rhee S-K (2020) Methylococcus geothermalis sp. nov., a methanotroph isolated from a geothermal field in the Republic of Korea. Int J Syst Evol Microbiol 70:5520–5530
Bodelier PL, Dedysh SN (2013) Microbiology of wetlands. Front Microbiol 4:79
Bowman J (2000) The methanotrophs - the families Methylococcaceae and Methylocystaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, Vol. http://link.springer-ny.com/link/service/books/10125. Springer Verlag, New York
Cervantes F, Garcia S, Peura S, Balagurusamy N (2021) Editorial: Methanotrophs: diversity, environmental relevance and applications. Front Microbiol 12(796861):1–3
Conrad R (2009) The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep 1:285–292
Costello AM, Lidstrom ME (1999) Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl Environ Microbiol 65:5066–5074
Cui J, Zhao J, Wang Z, Cao W, Zhang S, Liu J, Bao Z (2020) Diversity of active root-associated methanotrophs of three emergent plants in a eutrophic wetland in northern China. AMB Express 10:1–9
Dedysh SN (2009) Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies. Microbiol 78:655–669
Dedysh SN (2011) Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front Microbiol 2:184
Dedysh SN, Panikov NS, Liesack W, Grosskopf R, Zhou J, Tiedje JM (1998) Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science 282:281–284
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes - characterization of a gene coding for 16s-ribosomal Rna. Nucleic Acids Res 17:7843–7853
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Mol Biol Rev 60:439–471
Khalifa A, Lee CG, Ogiso T, Ueno C, Dianou D, Demachi T, Katayama A, Asakawa S (2015) Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere. Int J Syst Evol Microbiol 65:3527–3534
Khatri K, Mohite JA, Pandit PS, Bahulikar RA, Rahalkar MC (2019) Description of ‘Ca. Methylobacter oryzae’ KRF1, a novel species from the environmentally important Methylobacter clade 2. Antonie Van Leeuwenhoek 113:729–735
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Ogiso T, Ueno C, Dianou D, Huy TV, Katayama A, Kimura M, Asakawa S (2012) Methylomonas koyamae sp. nov., a type I methane-oxidizing bacterium from floodwater of a rice paddy field. Int J Syst Evol Microbiol 62:1832–1837
Pandit P, Rahalkar MC (2018) Renaming of ‘Candidatus Methylocucumis oryzae’ as Methylocucumis oryzae gen. nov., sp. nov., a novel Type I methanotroph isolated from India. Antonie Van Leeuwenhoek 112:955–959
Pandit PS, Hoppert M, Rahalkar MC (2018) Description of ‘Candidatus Methylocucumis oryzae’, a novel type I methanotroph with large cells and pale pink colour, isolated from an Indian rice field. Antonie Van Leeuwenhoek 111:2473–2484
Park S, Kim S-J (2019) Application and development of methanotrophs in environmental engineering. J Mater Cycles Waste Manag 21:415–422
Patil R, Mahajan M (2018) Herbaceous flora of weeds growing at ARAI hills. Int J Res Biosci Agric Technol 16:19
Rahalkar M, Khatri K, Pandit P, Bahulikar R, Mohite J (2021) Cultivation of important methanotrophs from Indian rice fields. Front Microbiol 12:1–15
Rahalkar MC, Khatri K, Mohite J, Pandit P, Bahulikar R (2020) A novel type I methanotroph Methylolobus aquaticus gen. nov. sp. nov. isolated from a tropical wetland. Antonie Van Leeuwenhoek 113:959–971
Saunois M, Stavert AR, Poulter B et al (2020) The global methane budget 2000–2017. Earth Syst Sci Data 12:1561–1623
Siljanen HM, Saari A, Bodrossy L, Martikainen PJ (2012) Seasonal variation in the function and diversity of methanotrophs in the littoral wetland of a boreal eutrophic lake. FEMS Microbiol Ecol 80:548–555
Strong PJ, Xie S, Clarke WP (2015) Methane as a resource: can the methanotrophs add value? Environ Sci Technol 49:4001–4018
Torres-Alvarado R, Fernández FJ, Barriga-Sosa I (2005) Methanogenesis and methane oxidation in wetlands: implications in the global carbon cycle. Hidrobiológica 15(3):327–349
Whittenbury R, Phillips KC, Wilkinson JF (1970) Enrichment, isolation and some properties of methane utilising bacteria. J Gen Microbiol 61:205–218
William G, Weisburg SMB, Pelletier DA, Lane DJ (1990) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Funding
This work was supported by the department of science and technology, SERB grants: (EMR/2017/002817), and POWER fellowship grant (SPF/2022/000045) provided to MCR. Author KP is thankful to the University Grant Commission for the junior research fellowship. Similarly, JAM acknowledges the SARTHI program for the junior research fellowship. SSM acknowledges SERB project: CRG/2021/000941 for providing for the junior research fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization: Monali C. Rahalkar and Rahul A. Bahulikar. Sample collection: Monali C. Rahalkar, Rahul Bahulikar, Jyoti Mohite, Shrinidhi Deshpande, Sanjana Patange, and Kajal Pardhi. Methodology: Jyoti A. Mohite, Shubha S. Manvi, Shrinidhi Deshpande, Sanjana Patange, Kajal Pardhi, Mansi Joshi, and Sharvari Kulkarni. Formal analysis and investigation: Jyoti A. Mohite, Kajal Pardhi, and Monali C. Rahalkar. Writing — initial draft preparation: Monali C. Rahalkar and Shubha Manvi. Writing — review and editing: Monali C. Rahalkar, Jyoti Mohite, Shubha Manvi, and Rahul A. Bahulikar. Funding acquisition: Monali C. Rahalkar. Supervision: Monali C. Rahalkar. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
There was no involvement of humans and/or animals in this study. And hence, the following declarations are not applicable.
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent to publish
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Accession numbers
pmoA: OQ354217, OQ354218, OQ354219, OQ354221, OR004530, OR004531, OR004532
16S rRNA gene: OQ373008, OQ373314, OQ373316, OQ373317
Supplementary information
ESM 1
(DOCX 1004 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mohite, J.A., Manvi, S.S., Pardhi, K. et al. Diverse type I and type II methanotrophs cultivated from an Indian freshwater wetland habitat. Int Microbiol 27, 607–614 (2024). https://doi.org/10.1007/s10123-023-00415-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-023-00415-4