Abstract
Background
To date, many efforts have been made to understand the resistance mechanism of trastuzumab in human epidermal growth factor receptor 2 (HER2)-positive breast and gastric cancer. However, there is still a huge unmet medical need for patients with trastuzumab resistance.
Methods
In our study, we generated four trastuzumab-resistant (HR) cancer cell lines from ERBB2-amplified gastric and biliary tract cancer cell lines (SNU-216, NCI-N87, SNU-2670, and SNU-2773).
Results
Here, we found higher PD-L1 expression in trastuzumab-resistant (HR) HER2-positive cancer cells than in parental cells, and blocking PD-L1 reversed the resistance to trastuzumab in HR cells. Trastuzumab upregulated PD-L1 expression via NF-κB activation in both parental and HR cells, however, led to DNA damage only in parental cells. The WEE1 inhibitor adavosertib, which downregulates PD-L1 expression, enhanced trastuzumab efficacy by blocking BRCA1-CMTM6-PD-L1 signals and the HER2-CDCP-1-SRC axis. Additionally, the levels of galectin-9, CD163, FoxP3, and CTLA-4 were diminished by blocking WEE1 in the presence of human PBMCs in vitro.
Conclusion
Taken together, the strategy of co-targeting HER2 and WEE1 could overcome resistance to trastuzumab in HER2-positive cancers, supporting further clinical development in HER2-positive cancer patients.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is common worldwide in terms of its incidence, and biliary tract cancer (BTC) is a relatively rare tumor that is more commonly diagnosed in Korea than in western countries [1, 2]. Notably, human epidermal growth factor receptor 2 (HER2) overexpression represents 10–15% and 10–20% of GC and BTC, respectively [3, 4]. As of 2019, several anti-HER2 targeted agents have been approved for patients with HER2-positive breast cancer and gastric cancer. Trastuzumab is a representative anti-HER2 monoclonal antibody [5] and, in combination with chemotherapy, is a standard of care for patients with HER2-positive GC and breast cancer. However, resistance remains a major challenge in HER2-targeted therapies.
Programmed cell death ligand 1 (PD-L1) is an immune checkpoint that plays a vital role in the tumor microenvironment and can affect the killing ability of immune cells [6]. Recent research has reported that trastuzumab could upregulate PD-L1 expression to evade the immune response in breast cancer cells [7, 8]. Furthermore, PD-L1 is involved in the DNA damage response (DDR) pathway of cancer cells [9]. Among the resistance mechanisms to trastuzumab, the role of PD-L1 modulation and its interaction with the DDR pathway has not yet been discovered in HER2-positive cancers.
DDR pathway alterations have been frequently found in patients with GC and BTC [10, 11]. Given the molecular characteristics, both GC and BTC are potential candidates for DDR-targeted drug development. WEE1 is a core member of the DDR signaling pathways [12,13,14]. We have evaluated the anti-tumor effect of a WEE1 inhibitor in pancreatic cancer [15] and reported that WEE1 inhibition dramatically downregulated PD-L1 expression in pancreatic cancer cells by reducing CKLF-like MARVEL Transmembrane Domain Containing 6 (CMTM6) expression.
In the current study, we aimed to evaluate the role of PD-L1 in trastuzumab resistance in HER2-positive cancers with or without immune cells and to explore WEE1 inhibition as a strategy to overcome trastuzumab resistance by PD-L1 modulation.
Materials and methods
Human cell lines and reagents
We established two patient-derived, ERBB2-amplified biliary tract cancer cell lines, SNU2670 and SNU2773, respectively [16]. SNU216 and NCI-N87, two ERBB2-amplified gastric cancer cell lines, were purchased from the Korean Cell Line Bank (Seoul, South Korea). All cells were cultured in RPMI-1640 medium (Welgene Inc., Gyeongsan, Korea) containing 10% fetal bovine serum and 10 µg/ml gentamicin at 37 °C with 5% CO2. Trastuzumab was purchased from Roche (Korea). The WEE1 inhibitor adavosertib was kindly provided by AstraZeneca (Macclesfield, Cheshire, UK). Trastuzumab-resistant (HR) cell lines were generated as previously described [17].
Cell viability assay
Cells were seeded in 96-well plates at a density of 0.5–8 × 103 cells per well, incubated overnight at 37 °C, and then exposed to various concentrations of trastuzumab and/or adavosertib for 72 h. No treatment was a control. A 50 µl aliquot of 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT) solution (Sigma-Aldrich, St. Louis, MO, USA) was added to each well and the incubation was continued at 37 °C for 4 h. The medium was removed and 150 µl of dimethyl sulfoxide (DMSO) was added to each well and mixed. The absorbance at 540 nm was measured with a VersaMax Microplate Reader (Molecular Devices, Sunnyvale, CA, USA). The experiments were performed three times.
Trypan blue staining
Cells (2 × 105) were seeded into 60-mm dishes and treated with 5 μg/ml of trastuzumab and/or 0.5 μM of adavosertib for 72 h. The cells were then stained with trypan blue solution (#T10282, Thermo Fisher Scientific, Waltham, MA, USA). Next, the cell viability was measured using Countess™ Automated Cell Counter (Thermo Fisher Scientific). The experiments were performed three times.
Colony-forming assay
Cells (0.5–7 × 103) were seeded in 6-well plates and exposed to various concentrations of trastuzumab for 10 days. The colonies were then stained with Coomassie Brilliant Blue for 2 h and counted using Gel Doc system software (Bio-Rad, Hercules, CA, USA). Each experiment was repeated three times.
SiRNA transfection
Cells (1.5–8 × 105) were seeded in 100-mm dishes and incubated 48 h with normal medium. SiRNAs specific for target genes and negative control were purchased from Genolution (Seoul, Korea). Cells were transfected with each siRNA at 50 nM final concentration for 24 h. Then cells were harvested and re-seeded for cell viability assay, colony-forming assay and western blotting. The sequence of specific siRNAs was used as follows. Negative control: 5′ CCUCGUGCCGUUCCAUCAGGUAGUU 3′, si-CD274 (PD-L1): 5′ GAAUCAACACAA- CAACUAAUU 3′, si-RELA (NF-κB p65): 5′ GACAAGUGGCCAUUGUGUUUU 3′, si-BRCA1: 5′ GGAAAUGGCUGAACUAGAAUU 3′, si-CREB1: 5′ GCAACCAAGUUGUU- GUUCAUU 3′, si-TEAD1: 5′ CAAACAGGGAUACACAAGAUU 3′. Each experiment was performed three times.
Western blot analysis
Cells (2–8 × 105) were seeded in 100-mm dishes and exposed at indicated conditions. The proteins were harvested as previously described [15]. Primary antibodies against the following molecules were purchased from Cell Signaling Technology (Beverley, MA, USA): WEE1 (#4936), p-WEE1-Ser642 (#4910), p-HER2-Tyr 1248 (#2247), HER2 (#2165), CDC2 (#9112), p-CDC2-Tyr15 (#9111), p-NF-κB p65-Ser536 (#3033), NF-κB p65 (#8242), CtIP (#9201), PD-L1 (#13684), p-SRC-Tyr 416 (#2101), SRC (#2108), p-CDCP-1-Tyr707 (#13111), CDCP-1 (#4115), BRCA1 (#9010), p-CREB-Ser133 (#4095), CREB (#4034), TEAD1 (#12292), FOXA2 (#3143), PCNA (#13110), CD163 (#93498), FOXP3 (#12632), CD8α (#85336). Anti-PD-1 antibody (#ab52587) was from Abcam Bioscience (Cambridge, UK); Anti-β-actin antibody was from Sigma-Aldrich; anti-γH2AX antibody (#05-636) was from Millipore (Billerica, MA, USA); anti-CTLA-4 (#sc-9094) and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; #sc-25778) were from Santa Cruz Biotechnology (Dallas, TX, USA); anti-CMTM-6 (#PA5-34747), Galectin-9 (#PA5-50966), and secondary antibodies were from Thermo Fisher Scientific.
Cell cycle analysis
Cells (2 × 105) were seeded in 60-mm dishes and treated with adavosertib (0, 0.25, and 0.5 μM) for 48 h. The cells were then harvested and fixed with 70% ethanol at − 20 °C for 2 days. An aliquot of 7 µl of 20 mg/ml RNase A (Invitrogen, Carlsbad, CA, USA) was added to each well and the plates were incubated for 10 min at 37 °C. Finally, propidium iodide (PI; Sigma-Aldrich) was added to each well and the cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Each experiment was repeated three times.
Immunoprecipitation
Cells were seeded in 150-mm dishes at a density of 1 × 106 cells/dish, treated with adavosertib for 72 h. Anti-BRCA1 antibody (#9010, Cell Signaling Technology), anti-IgG antibody (#2729, Cell Signaling Technology) and Protein A/G PLUS agarose (#sc-2003, Santa Cruz Biotechnology) were used. Finally, samples were analyzed by western blotting as described above. Each experiment was performed three times.
Comet assay
Cells (2–4 × 105) were treated with 5 μg/ml of trastuzumab and/or 0.5 μM of adavosertib for 72 h. Then alkaline comet assay was conducted as previously described [15]. Tail moment (migration of DNA fragments) and intensity (DNA content) were measured using the Comet Assay IV program (Andor Technology, Belfast, UK). Each condition was analyzed in three independent experiments.
Human cytokine array
Cells (2–4 × 105) were seeded in 60-mm dishes and exposed to trastuzumab and/or adavosertib for 24 h. The cell supernatant was then collected and 700 μl of each sample was analyzed using the Proteome Profiler Human Cytokine Array Kit (#ARY005B, R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. Spot intensities were measured using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Immunofluorescence
For immunofluorescence, cells (1.5–2 × 105) were seeded into confocal dishes. After 24 h, trastuzumab (5 μg/ml) and/or adavosertib 0.5 μM were added for 48 h. Subsequently, the cells were fixed by 4% paraformaldehyde (#P2031, Biosesang, Gyeonggi-do, Korea) for 10 min and then permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 5 min. The primary antibodies were used as follows. Anti-PD-L1 (#59949, Cell Signaling; 1:50), anti-CMTM6 (#PA5-55472, Thermo Fisher; 1:100). Goat anti-Rabbit IgG Antibody Alexa Fluor 594 (#11012, Thermo Fisher; 1:100) and Alexa Fluor 488 (#11008, Thermo Fisher; 1:100) were used as secondary antibodies. The cells were stained with DAPI solution (Sigma-Aldrich) for 1 min. The image was captured using a Zeiss LSM 510 laser scanning microscope (Carl Zeiss). The experiment was repeated three times. The signal intensity was analyzed by ImageJ software.
In vitro human PBMC-mediated killing assay
Primary peripheral blood mononuclear cells (PBMC) were obtained from healthy donors according to the Institutional Review Board of Seoul National University Hospital guidelines (approval number: H-1811-106-987). The isolated PBMC was activated with 100 ng/ml of anti-CD3 antibody (#160037-85, Invitrogen; Waltham, MA, USA) and human recombinant Interleukin-2 (#78036.3, STEMCELL Technologies; Vancouver, Canada) for 3 days. Cancer cells were labeled with CellTrace™ Far Red Cell Proliferation Kit (#34564, Invitrogen) and seeded in 60-mm dishes at a density of 2–4 × 105 cells per dish. After 24 h of incubation, activated PBMC (Cancer cell: PBMC = 1:10) and CD3/CD28 tetrameric antibody complexes were added (#10991, STEMCELL Technologies), followed by trastuzumab (5 μg/ml) and/or adavosertib (0.5 μM) treatment for 48 h. Then the cancer cells and PBMC were harvested and permeabilized and washed using Fixation/Permeabilization Solution Kit (#554714, BD Bioscience, Franklin Lakes, NJ, USA). Finally, the percentage of V450-cleaved caspase 3 (#560627, BD Biosciences) was detected by BD FACS Canto II system. The experiment was repeated three times.
Mouse xenograft experiments
Animal experiments were performed at the Biomedical Center for Animal Resource Development of Seoul National University (Seoul, Korea) according to institutional guidelines, and prior approval of the study protocol was obtained from the Institutional Animal Care and Use Committee. Four-week-old female athymic nude mice were purchased from Orient Bio Inc. (Gyeonggi-do, South Korea). SNU2670HR cells were resuspended at 2 × 107 cells in 100 μl of PBS and injected subcutaneously. The tumor volume was calculated using the formula: volume = [(width)2 × height]/2. When the tumor volume reached 500 mm3, the mice were randomly assigned to four groups of four mice to receive (1) vehicle (2-hydroxypropyl-β-cyclodextrin solution), (2) trastuzumab was injected intraperitoneally twice weekly at 4 mg/kg for 3 weeks, or (3) adavosertib was administered by oral gavage once daily at 60 mg/kg for 3 weeks (5 days on/2 days off), (4) combination treatment for 3 weeks. Body weights and tumor sizes were measured every other day.
Immunohistochemistry
Sections of paraffin-embedded xenograft tumor tissues were deparaffinized and dehydrated. Proliferating cells were detected by staining with anti-Ki-67 antibody (GeneTex Inc.) at a dilution of 1:100. Anti-PD-L1 (#13684, cell signaling), anti-BRCA1 (#ab16780, Abcam biosciences), and anti-CMTM6 antibodies (#PA5-34747, Thermo Fisher Scientific) were used at dilutions of 1:200.
Mouse cytokine array
Immediately before killing, the mice were bled and serum and spleen samples were prepared. Aliquots of 500 μl of serum and 300 μg of spleen lysates were analyzed using the Proteome Profiler Mouse XL Cytokine Array Kit (#ARY028, R&D Systems) according to the manufacturer’s instructions. Spot intensities were measured using ImageJ software.
Statistical analysis
Analyses were conducted using SigmaPlot version 10.0 (Systat Software Inc., San Jose, CA, USA). Data are presented as the means ± standard errors (SE). All statistical tests were two sided. Differences were considered significant if the p values were < 0.05. Combined drug effects were analyzed by calculating the combination index (CI) with CalcuSyn software (Biosoft, Cambridge, United Kingdom). CI values of < 1, 1, and > 1 indicate synergistic, additive, and antagonistic effects, respectively.
Results
Knockdown of PD-L1 enhances the anti-proliferative effects of trastuzumab in HR cells
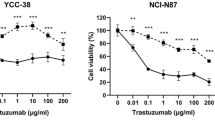
To understand the acquired resistance mechanisms to trastuzumab, the characteristics of HR cells were evaluated. First, we detected the basal protein expression levels to compare the difference between parental and HR cells (Fig. 1a). PD-L1 expression was upregulated in all HR cells compared with their parent cells.
Characteristics of trastuzumab-resistant (HR) cells. a Four HR cell lines were generated, and the basal expression of PD-L1 was detected using western blotting. The data represent three independent experiments. b Cells were transfected with PD-L1-specific siRNAs or control siRNAs for 24 h. Next, the MTT assay was conducted using different concentrations of trastuzumab (0, 0.01, 0.1, 1, 10, and 100 μg/ml) for 72 h. PD-L1 expression was detected using western blotting after the MTT assay. All the experiments were repeated three times. *p < 0.05, **p < 0.01. c and d Cells were transfected with PD-L1-specific siRNAs or control siRNAs for 24 h. Subsequently, the cells were re-harvested for the colony-forming assay. Cells were exposed to dose-dependent trastuzumab (0, 0.1, 1, 10, and 100 μg/ml) for 10 days. All the experiments were repeated three times. *p < 0.05, **p < 0.01, ***p < 0.001
PD-L1 expression was disrupted using small interfering RNAs (siRNAs) in all cell lines, and then the MTT assay and colony-forming assay were performed (Fig. 1b, c). As determined by the MTT assay, no significant difference was found in the response of the parental cells to trastuzumab treatment with PD-L1 knock-down. By contrast, under transfection with si-PD-L1, growth was inhibited in HR cells with trastuzumab treatment to the same extent as that observed in parental cells (Fig. 1b). To confirm the anti-proliferative effect of si-PD-L1 with trastuzumab treatment, the colony-forming assay was conducted. As shown in Fig. 1c, knock-down of PD-L1 in all cell lines led to reduced colony formation. Additionally, the effect was more profound in HR cells than in parent cells (Fig. 1d). These data suggested that upregulated PD-L1 expression might partially contribute to resistance to trastuzumab in HR cells.
Trastuzumab mediates nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation, which contributes to PD-L1 upregulation
To identify the mechanism of PD-L1 upregulation by trastuzumab in HER2-positive cancer cells, the signals related to PD-L1 expression were assessed by western blotting. All four HR cell lines exhibited upregulated phosphorylation of NF-κB p65 compared with parental cells (Fig. 2a). Given that NF-κB activation has been identified as a transcriptional enhancer of PD-L1 expression previously [18], we asked whether the increased p-NF-κB levels contributed to the PD-L1 upregulation in HR cells. Subsequently, we randomly selected one of the HR cell lines, SNU2670HR, and then transfected the cells with si-RELA before trastuzumab treatment. Treatment with 5 μg/ml of trastuzumab for 24 h led to PD-L1 upregulation in the control group but not in the NF-κB p65 knockdown group (Fig. 2b). Additionally, we observed that trastuzumab induced the accumulation of γ-H2AX, a DNA damage marker, only in parental cells (Fig. 2c). Here, we wanted to evaluate whether DDR pathway is related to PD-L1 expression.
Mechanism of PD-L1 upregulation by trastuzumab. a Western blotting was performed to detect PD-L1-related proteins. The data represent three independent experiments. b Cells were transfected with RELA-specific siRNAs or control siRNAs for 24 h. Next, SNU2670HR cells were treated with 5 μg/ml of trastuzumab for 24 h. PD-L1 expression was evaluated by western blotting. All the experiments were repeated three times. c The indicated cell lines (NCI-N87/NCI-N87HR/SNU2670/SNU2670HR) were exposed to 5 μg/ml of trastuzumab for 24 h. The proteins were then isolated to conduct western blotting. Each experiment was repeated three times
WEE1 inhibition reduces PD-L1 expression by BRCA1 downregulation
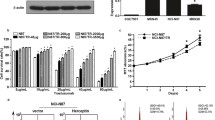
In line with our previous findings [15], Fig. 3a displays that WEE1 inhibition reduced PD-L1 expression in a dose-dependent manner in both parental and HR cells. Next, we selected two groups of cells to conduct subsequent experiments, NCI-N87/NCI-N87HR (GC) and SNU2670/SNU2670HR (BTC). Interestingly, the population of sub-G1 was significantly increased by WEE1 inhibition, especially in HR cells (Fig. 3b). To better understand the anti-tumor effects of the WEE1 inhibitor, the cells were exposed at different times of adavosertib treatment. Consistent with the above data, PD-L1 expression was disrupted by adavosertib within 48 h, accompanied by CMTM6 reduction (Fig. 3c). Moreover, we detected the downregulation of p-SRC, phosphor-CUB domain-containing protein 1 (p-CDCP-1), and p-HER2 levels in all four cell lines. Additionally, in response to adavosertib, all the cell lines displayed a dramatic reduction of BRCA1, forkhead box protein A2 (FOXA2), and phosphor-CAMP-responsive element binding protein (p-CREB) expression, but TEA domain family member 1(TEAD-1) expression remained unchanged in SNU2670 cells (Fig. 3c). TEAD-1 is a transcription factor that has been shown to promote tumorigenesis in GC and BTC [19, 20]. Based on our cell cycle analysis, SNU2670 is less sensitive to trastuzumab than the other three cell lines. Whether unaltered TEAD-1 expression is related to less sensitivity to adavosertib remains to be further evaluated (Fig. 3b). These data above strongly suggest that adavosertib can efficiently block the transcription factors involved in the DDR signaling pathway.
Anti-tumor effect of a WEE1 inhibitor (adavosertib) in HER2-positive GC and BTC cells. a Cells were treated with different doses of adavosertib (0, 0.25, and 0.5 μM) for 72 h. Western blotting was conducted three times. b The indicated cell lines (NCI-N87/NCI-N87HR/SNU2670/SNU2670HR) were treated with different doses of adavosertib (0, 0.25, and 0.5 μM) for 48 h to perform cell cycle analysis. The sub-G1 population with fold change was analyzed using three independent datasets. *p < 0.05, **p < 0.01. c The indicated cell lines (NCI-N87/NCI-N87HR/SNU2670/SNU2670HR) were exposed to 0.5 μM adavosertib in a time-dependent manner (0, 12, 24, 48, and 72 h). Various proteins were evaluated by western blotting. All the experiments were repeated three times. d Western blotting was performed on SNU2670HR cells left untreated or treated with 0.5 μM adavosertib, 1 μM MG132, or their combination for 48 h. The data represent three independent experiments. e–h SNU2670HR cells were transfected with BRCA1-specific siRNAs, CREB1-specific siRNAs, TEAD-1-specific siRNAs, PD-L1-specific siRNAs, or control siRNAs for 24 h. The related molecules were detected by western blot analysis. All the experiments were repeated three times. g SNU2670HR cells were left untreated or were treated with 0.5 μM adavosertib. After 72 h, immunoprecipitation and western blotting were performed to analyze the interaction between BRCA1 and WEE1. All the experiments were repeated three times
Recently, CMTM6 has been identified as a PD-L1 post-transcriptional regulator [21]. To evaluate whether CMTM6 reduction is correlated with PD-L1 expression in our study, SNU2670HR cells were exposed to adavosertib with or without the proteasome inhibitor MG132 for 48 h (Fig. 3d). Adavosertib did not downregulate PD-L1 expression in the presence of MG132. Next, we asked how the WEE1 inhibitor mediates CMTM6 expression in HER2-positive cells. Based on our findings above, we speculated that CMTM6 might be mediated by WEE1 inhibition via transcription factors such as BRCA1, CREB, or TEAD-1. To confirm this hypothesis, the expression levels of both PD-L1 and CMTM6 were detected after transfection with si-BRCA1, si-CREB, or si-TEAD-1 in SNU2670HR cells (Fig. 3e). Interestingly, we observed that all the constructs contributed to decreased expression of PD-L1 and CMTM6, but si-BRCA1 blocked the expression of PD-L1 more profoundly than si-CREB or si-TEAD-1. We conclude that WEE1 inhibition mediates CMTM6 expression through multiple signals. Most recently, inconsistent with our data, PD-L1 has been demonstrated to be an upstream signal of BRCA1 in the DDR pathway [9]. Therefore, we knocked down PD-L1 expression using si-PD-L1 in SNU2670HR cells, and the BRCA1 expression level was evaluated. However, we detected no change in BRCA1 expression (Fig. 3f). Additionally, we performed immunoprecipitation to address the interaction between BRCA1 and WEE1. Notably, the levels of WEE1 bound to BRCA1 were dramatically reduced in the presence of adavosertib (Fig. 3g). Based on our observations, we hypothesized that the CMTM6-PD-L1 signaling pathway might be regulated by BRCA1 after treatment with adavosertib.
Furthermore, we tried to understand the mechanism of reduction of p-HER2/p-SRC/p-CDCP1 signals by adavosertib. The interaction among HER2, SRC, and CDCP-1 has been reported in breast cancer cells previously [22]. HER2-SRC crosstalk depends on the expression levels of p-CDCP-1 on the cell surface. Adavosertib-mediated p-CDCP1 reduction may have contributed to the downregulation of the p-HER2 and p-SRC levels (Fig. 3c). Here, when BRCA1 or CREB-1 expression was disrupted in SNU2670HR cells, the p-CDCP1 level was significantly declined (Fig. 3h). These results prompted us to perform a combination treatment of trastuzumab and adavosertib in all parental and HR cell lines.
WEE1 inhibition augments trastuzumab efficacy in HR cells
To assess the combined anti-proliferative effects, the MTT assay was conducted for 72 h. The combination index (CI) value at ED50 illustrated synergism in all HR and some parental cells, but not in NCI-N87 cells (Fig. 4a). Next, we utilized Trypan blue staining to monitor cell viability under the different treatments in SNU2670HR cells. As expected, trastuzumab (93%) did not affect the cell viability compared with the control (91%), adavosertib (63%) remarkably reduced the cell viability, and combination treatment (33%) had more potent efficacy than each drug alone (Fig. 4b). The comet assay data also supported the rationale for the combination strategy in HR cells. Trastuzumab-induced DNA damage occurred in NCI-N87 and SNU2670 cells, but not in NCI-N87HR and SNU2670HR cells (Fig. 4c). However, analysis of the tail intensity and moment demonstrated a synergistic combination effect in both parental and HR cells. To confirm, we monitored several representative signals related to cell proliferation and DDR using NCI-N87/HR and SNU2670/HR cell lines (Fig. 4d). HER2 phosphorylation was strongly reduced in co-treated cells compared with cells treated with each drug alone. The combination treatment also remarkably blocked p-CDC2 expression in both parental and HR cells. Additionally, trastuzumab-induced PD-L1 upregulation was downregulated by adavosertib. CMTM6 expression was not interrupted by trastuzumab but was decreased by adavosertib. The CtBP (carboxy-terminal binding protein) interacting protein (CtIP), a DNA double-strand break (DSB) resection factor, was downregulated in adavosertib and co-treated cells, but more obviously in HR cells [23]. Proliferating cell nuclear antigen (PCNA) and p-SRC were more dramatically decreased in HR cells than in parental cells by adavosertib /combination treatment. Next, immunofluorescence assay was performed using SNU2670 and SNU2670HR cells (Fig. 4e). Analysis of images showed that trastuzumab significantly increased PD-L1 expression in both parental and HR cells, and dual treatment with trastuzumab and adavosertib synergistically decreased the PD-L1 level. In line with the western blotting data, CMTM6 expression was efficiently reduced after combination treatment but was not regulated by trastuzumab alone. Because cytokines play important roles in the tumor microenvironment [24], we measured the cytokine release levels in SNU2670 and SNU2670HR cells (Fig. 4f). Notably, SNU2670 cells released very low levels of cytokines that were barely detectable within 24 h. However, SNU2670HR cells released several cytokines, including chemokine (C-X-C motif) ligand 1 (CXCL-1), interleukin-6 (IL-6), IL-8, macrophage migration inhibitory factor, and plasminogen activator inhibitor-1 (PAI-1). Among them, the CXCL-1 and PAI-1 levels were reduced by trastuzumab or adavosertib alone but were more significantly decreased under the combination conditions. These data suggest that increased cytokine release might partially contribute to trastuzumab resistance, and combination therapy could diminish cytokine release.
Combination effects of trastuzumab and adavosertib. a Different concentrations of trastuzumab (0, 0.01, 0.1, 1, 10, and 100 μg/ml), adavosertib (0, 0.001, 0.01, 0.1, 1, and 10 μM), and trastuzumab plus adavosertib were added to SNU2670HR cells for 72 h. Cell viability was calculated using the MTT assay, and the combination index at ED50 was analyzed using CalcuSyn software. All the experiments were repeated three times. CI values of < 1, 1, and > 1 indicate synergistic, additive, and antagonistic effects, respectively. b Three days after the addition of trastuzumab alone (5 μg/ml), adavosertib alone (0.5 μM), or their combination, the cells were stained with Trypan blue and then the cell viability was monitored. Green indicates live cells, and red indicates dead cells. The data represent three independent experiments. c Cells were incubated with the indicated doses of trastuzumab (5 μg/ml), adavosertib (0.5 μM), or their combination for 72 h. The comet assay was performed three times, and the tail intensity and moment were analyzed. *p < 0.05, **p < 0.01, ***p < 0.001. d Western blotting was conducted after treatment with trastuzumab (5 μg/ml), adavosertib (0.5 μM), or their combination for 72 h. The data represent three independent experiments. e Representative immunofluorescence images of the indicated cells after treatment with trastuzumab (5 μg/ml), adavosertib (0.5 μM), or their combination for 48 h. Blue indicates DAPI, red indicates PD-L1, and green indicates CMTM6. The intensity was measured using ImageJ. The data represent three independent experiments. *p < 0.05, ***p < 0.001, ns: no significance. f Human cytokine array analysis of the indicated cells treated with trastuzumab (5 μg/ml), adavosertib (0.5 μM), or their combination for 24 h. Spot intensities were quantified using ImageJ software. 1, CXCL-1; 2, IL-6; 3, IL-8; 4, MIF; 5, PAI-1. *p < 0.05, **p < 0.01, ***p < 0.001
WEE1 inhibition reverses trastuzumab resistance in the presence of PBMCs
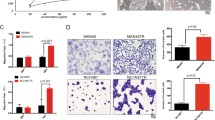
To better evaluate the anti-tumor effects of the combination strategy, SNU2670 or SNU2670HR cells were co-cultured with or without human peripheral blood mononuclear cells (PBMCs). As shown in Fig. 5a, human PBMCs were activated by IL-2 and CD3 for 72 h and then were co-incubated with cancer cells (cancer cells:PBMCs = 1:10) with or without drug treatment for 48 h. First, we detected the cleaved caspase-3 population to evaluate whether the combination strategy modulated PBMC-mediated killing ability. In SNU2670 cells, both trastuzumab and adavosertib significantly induced cleaved caspase-3 increase, and this effect was enhanced under PBMC co-cultured conditions (Fig. 5b). In SNU2670HR cells, trastuzumab did not affect caspase-3 cleavage while adavosertib showed dramatically increased caspase-3 cleavage. In the presence of PBMCs, trastuzumab reduced caspase-3 cleavage compared with control cells, but adavosertib and combination treatment still showed excellent killing ability versus the control group (Fig. 5b). Second, to better understand the effects of trastuzumab and adavosertib on immune modulation, we performed western blotting to monitor the signal changes of several immune parameters with or without PBMCs (Fig. 5c). Based on our observation, galectin-9 was downregulated by adavosertib in both SNU2670 and SNU2670HR cells but was more blocked by combination treatment in the presence of PBMCs. CD163, an M2-type macrophage marker, was also reduced by adavosertib or combined treatment. In addition, trastuzumab increased the expression of FoxP3 and CTLA-4, which are mainly expressed on regulatory T (Treg) cells, whereas adavosertib reduced this expression. Consistent with the previous results, PD-L1 was upregulated after trastuzumab treatment in both SNU2670 and SNU2670HR cells in the presence or absence of PBMCs. Although strong expression of PD-L1 was detected in PBMCs mixed with SNU2670 or SNU2670HR cells, adavosertib reduced PD-L1 expression dramatically. Given that PD-L1 is expressed on cancer cells and immune cells, we speculated that adavosertib might also block PD-L1 expression in immune cells. However, we detected no change in PD-1 expression after trastuzumab or adavosertib exposure. Importantly, in response to adavosertib, CD8 expression was enhanced in both parental and HR cells. Here, according to the above results, we realized that WEE1 inhibition could overcome trastuzumab resistance in HER2-positive cancer cells in vitro, even in the presence of PBMCs.
Combination effects of trastuzumab and adavosertib in the presence of human PBMCs. a Schematic diagram for the in vitro procedures of the human PBMC killing assay. b Cells were exposed to trastuzumab (5 μg/ml), adavosertib (0.5 μM), or their combination for 48 h in the absence or presence of activated human PBMCs. Next, the cleaved caspase-3 percentage was detected by flow cytometry. The experiments were repeated three times. *p < 0.05, **p < 0.01, ***p < 0.001. c The cells were treated as shown (b). The related immune parameters were captured by western blotting. The data represent three independent experiments
Co-targeting of HER2 and WEE1 shows strong anti-tumor growth activity in vivo
To confirm the anti-tumor activity of the combination strategy, we established a mouse xenograft model using SNU2670HR cells. After three weeks of treatment, trastuzumab alone did not affect tumor growth (Fig. 6a). Adavosertib monotherapy significantly delayed tumor growth, and the combination strategy showed a stronger inhibitory effect on tumor growth than each drug alone (Fig. 6a). During the animal experiments, none of the animals showed significant body weight loss (Fig. 6b). Regarding immunohistochemistry staining of tumor tissues and consistent with our in vitro findings, PD-L1 expression was increased by trastuzumab treatment and was reduced by adavosertib exposure (Fig. 6c). High levels of CMTM6 and BRCA1 expression were detected in both control and trastuzumab-treated groups but were strongly downregulated in the adavosertib -alone and combination groups. Expression of the proliferation marker ki67 was reduced by adavosertib and combination treatment but remained unchanged by trastuzumab monotreatment. Moreover, western blotting from isolated tumors showed that the combination therapy had strong inhibitory effects on tumor growth signals in HR xenograft models (Fig. 6d). The expression of p-HER2 was blocked by trastuzumab and further downregulated by additional treatment with adavosertib. The WEE1 inhibitor adavosertib (60 mg/kg) efficiently blocked p-WEE1 expression. We also confirmed that trastuzumab upregulated PD-L1 expression, while adavosertib downregulated PD-L1 expression. Similar to the IHC results, BRCA1 expression was attenuated in the adavosertib mono-and combination groups. Importantly, beyond PD-L1, another immune checkpoint molecule, galectin-9, was dramatically blocked by combination treatment. As expected, adavosertib alone and combination treatment increased the γ-H2AX level, which represents DNA damage accumulation. We also found that CD163 expression was reduced by adavosertib alone and combination treatment, therefore, we speculated that adavosertib might change tumor microenvironment to some extent through reverse M2 type macrophage polarization (Fig. 6d).
Anti-tumor effects of combination therapy in the SNU2670HR xenograft mouse model. a SNU2670HR tumor growth curves of the control, trastuzumab (4 mg/kg), adavosertib (60 mg/kg), and combination groups. *p < 0.05, **p < 0.01, ***p < 0.001. b Body weights (g) in SNU2670HR-xenografted mice within days after treatment. (c) The pathologic examinations were conducted using hematoxylin and eosin (H&E) staining (× 400). Immunohistochemical analysis of PD-L1 (× 400), CMTM6 (× 400), BRCA1 (× 400), and Ki67 (× 400) staining in SNU2670HR xenograft-excised tumors. d Western blot analysis of various proteins harvested from isolated tumors. The data represent three independent experiments. e Mouse cytokine array analysis of serum and spleens collected from xenografted mice. Spot intensities were measured using ImageJ software. 1. CCL6; 2. IGFBP-2; 3. Reg3G; 4. YKL-40; 5. CD105; 6. MMP9. *p < 0.05, **p < 0.01, ***p < 0.001
Finally, we performed mouse cytokine release analysis using mouse sera and spleens. As shown in Fig. 6e, among the various cytokines, the chemokine (C–C motif) ligand 6 (CCL6), insulin-like growth factor-binding protein 2 (IGFBP-2), and regenerating islet-derived protein 3 gamma (Reg3G) levels were synergistically reduced by combination treatment in the serum samples. The cytokine production by spleens was also measured subsequently. Dual combination therapy remarkably reduced the chitinase-3-like protein 1 (YKL-40), CD105, and matrix metallopeptidase 9 (MMP-9) levels compared with monotherapy (Fig. 6e). Given that these cytokines have been reported to be related to tumor progression, the regulatory mechanism of trastuzumab and adavosertib in cytokine release in HR cells needs further investigation [25, 26]. In summary, WEE1 inhibition overcomes trastuzumab resistance in HR tumor-bearing mice.
Discussion
To better identify the mechanism of HER2 resistance, continuous efforts have been underway in the past decade [5, 27]. Previously, trastuzumab has been shown to upregulate PD-L1 expression in breast cancer [7, 8]. In those studies, trastuzumab treatment upregulated PD-L1 expression in HER2-positive breast cancer cells under PBMC co-incubated conditions, leading to trastuzumab resistance [7]. In contrast to our data, they did not observe PD-L1 upregulation without PBMC environment. Notably, another study demonstrated that trastuzumab increased the PD-L1 expression in tumor-associated macrophages in both HER2-positive breast cancer patients and a humanized mouse model [8]. Although we did not specifically detect PD-L1 expression in macrophages, based on Fig. 5c data, trastuzumab enhanced PD-L1 expression in the presence of PBMCs. We speculated that trastuzumab might induce PD-L1 upregulation in both tumor cells and immune cells in HER2-positive cancer cells. The most recent data revealed that the dual inhibition of HER2 and PD-L1 successfully enhanced the anti-tumor effect of anti-HER2 therapy alone in a mouse model [28]. Importantly, pembrolizumab (PD-1 inhibitor) in combination with trastuzumab therapy exhibited an objective response in patients with HER2 (+) PD-L1 (+) breast cancer in a phase Ib-II clinical trial [29]. A phase III clinical trial is ongoing for patients with HER2-positive advanced GC and gastroesophageal junction adenocarcinoma using the combination of pembrolizumab plus trastuzumab plus chemotherapy (ClinicalTrial.gov, NCT03615326). Our study provides a rationale that the PD-1/PD-L1 axis can affect the anti-tumor effect of trastuzumab in HER2-positive cancers. In our study, we demonstrated that high PD-L1 expression in HR cells contributed to trastuzumab resistance and could be overcome by WEE1 inhibition through BRCA1 reduction.
Because PD-L1 plays a pivotal role in the immune response [30], the PD-L1 regulatory mechanisms have been uncovered. Regarding the transcriptional levels, hypoxia-inducible factor alpha, signal transducer and activator of transcription 3, NF-ĸB, and Myc have been identified as regulators of the CD274 mRNA level [31, 32]. Concerning the post-transcriptional levels, CMTM6, glycogen synthase kinase 3, dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit STT3, and cyclin-dependent kinase 4 contribute to the stabilization of PD-L1 expression in multiple cancer types [33]. In our previous publication, we observed that the wee1 inhibitor (adavosertib) reduced PD-L1 and CMTM6 expression in pancreatic cancer cells [15]. Therefore, we focused on revealing the regulatory mechanisms of WEE1 inhibitors on CMTM6-PD-L1. Consistent with our previous finding, adavosertib dramatically downregulated the CMTM6-PD-L1 signaling pathway in HER2-positive cancer cells, and we observed that BRCA1/CREB/TEAD1 were involved in CMTM6 modulation.
To our knowledge, CMTM6 is a novel protein whose function has been rarely understood thus far. Most recently, Guan and colleagues found that high expression of CMTM6 in gliomas is associated with a poor survival rate and positively regulates T-cell tolerance and Treg-cell differentiation [34]. In non-small cell lung cancer, CMTM6 expression was detected in approximately 70% of cases, and co-expression of CMTM6 and PD-L1 in the stroma was correlated with longer overall survival in patients receiving immunotherapy [35]. However, no evidence exists currently on the role of CMTM6 expression in GC or BTC. Notably, the regulatory mechanism of CMTM6 expression has not been studied in any type of cancer.
Several immune cell markers were assessed by trastuzumab or adavosertib treatment using healthy donor PBMCs in our study (Fig. 5). One of our major novel findings was that CD163 (M2 type macrophage), FoxP3 (Treg), and CTLA-4 (Treg) were downregulated by adavosertib in both parental and HR cells. We also monitored the reduction of galectin-9 expression by the combination strategy in both in vitro and in vivo models. As a T-cell immunoglobulin and mucin-domain containing-3 partner, high levels of galectin-9 negatively modulate the immune response and are associated with a poor prognosis in patients with solid tumors [36, 37]. The association between T-cell immunoglobulin and mucin-domain containing-3/galectin-9 and WEE1 has not been assessed previously. Concerning the role of galectin-9 in the tumor microenvironment, our finding provided more detailed evidence for the use of the WEE1 inhibitor. In addition, as shown in Fig. 4f, adavosertib reduced PAI-1 release in HR cells, interrupting M2 macrophage recruitment and polarization [38]. Our data supported that adavosertib might modulate macrophage polarization and the Treg-cell population in tumor microenvironment to enhance the immune response. In xenograft experiments, the release of several mouse cytokines was significantly suppressed by combination therapy, including CCL6, IGFBP-2, Reg3G, YKL-40, CD105, and MMP-9. High CCL6 levels promote fibroblast differentiation and Treg cell function, leading to immune suppression in the pancreatic cancer tumor microenvironment [39]. In solid tumors, overexpression of IGFBP-2, Reg3G, YKL-40, and MMP-9 is associated with tumorigenesis through accelerating immunosuppression [25, 26, 40]. The depletion of CD105 is also involved in DNA repair disruption in prostate cancer [41]. Although the role of adavosertib in cytokine release is incompletely understood, we highly suggest that the immune response may be enhanced by adavosertib treatment.
Furthermore, earlier studies described that adavosertib modulates immune parameters to enhance the immune response [42, 43]. For example, human head and neck cancer cells responded well to natural killer (NK) cell therapy in the presence of adavosertib, and a similar result was observed in murine oral cells [42]. The modulation of NK cell function mediated by adavosertib is very expected since one of the modes of action of trastuzumab is antibody-dependent cellular cytotoxicity (ADCC). When NK cells are activated, they release a large number of cytokines such as interferon, which leads to an increase in the expression of PD-L1 in tumor cells and further inhibits the effect of NK cells [44]. Effector NK cells are also negatively affected by galectin-9 level [45]. As shown in Fig. 5c, adavosertib downregulated both PD-L1 and galectin-9 expression in both SNU2670 and SNU2670HR cells. These findings support WEE1 inhibition might enhance trastuzumab-mediated ADCC effect.
Another research team has demonstrated that G2/M cell cycle checkpoint activation is a resistance mechanism of cytotoxic T lymphocyte (CTL) killing ability and could be reversed by adavosertib [43]. In the present study, CD8α expression was enhanced after adavosertib treatment in the presence of human PBMCs (Fig. 5c). Because CD8α is predominantly expressed on CTL but is also expressed on dendritic cells and natural killer cells, further study is needed to monitor which CD8α in immune cells is modulated by adavosertib.
As a potent DDR-targeted agent, adavosertib has been studied in different cancer types in both preclinical and clinical trials [46]. Consistent with previous publications, adavosertib significantly suppressed cell proliferation and cytokine release, increased the sub-G1 population and DNA double-strand breaks, and blocked related signals. Here, we further discovered that adavosertib could diminish the p-HER/p-CDCP-1/p-SRC signaling pathway in both parental and HR cells through direct blockade of BRCA1 and p-CREB expression (Fig. 3c, h). Interestingly, upregulated CDCP-1 has been shown to stimulate SRC activation, leading to trastuzumab resistance in HER2-positive breast cancer [22]. Moreover, we have previously reported that SRC activation partially contributes to trastuzumab resistance in HER2-positive GC and BTC cells [17].
PD-L1 inhibition might reverse trastuzumab resistance in HER2-positive cancer cells. The WEE1 inhibitor has potent anti-tumor effects through inducing apoptosis, DNA strand breaks, and inhibition of cell growth while downregulating PD-L1. Previously, trastuzumab combined with a PARP inhibitor (olaparib) showed more potent anti-tumor effects than trastuzumab alone in HER2-positive breast cancer [47]. Another research team has demonstrated that NF-κB -IL8 signaling pathway is the main trastuzumab resistance mechanism and PARP inhibitor could block the PARP1- NF-κB -IL8 signals to overcome trastuzumab resistance in HER2-positive breast cancer [48]. In our study, si-PD-L1 can reverse trastuzumab resistance in HR cells, and WEE1 inhibitor not only downregulates PD-L1 expression but also increases PARP cleavage (data not shown), but does not affect IL-8 level. Our data supported that NF-κB -PD-L1 upregulation might be a core resistance mechanism at least in gastric and biliary tract cancer. Regarding PARP inhibitors, PD-L1 expression can be upregulated by PARP inhibitors such as olaparib [49]. Therefore, we speculated that the WEE1 inhibitor might be more suitable than the PARP inhibitor to overcome trastuzumab resistance. In addition, the HER2 expression level was detected in our previous study. HER2 level is upregulated in two HR cell lines (SNU216HR and NCI-N87HR) compared to respective parental cells and is reduced in the other two HR cell lines (SNU2773HR and SNU2670HR) [17]. HER2 loss might be one of trastuzumab resistance mechanisms in some HR cells, but not all cells. Given these circumstances, using the WEE1 inhibitor against trastuzumab resistance is recommended, especially in patients with post-trastuzumab treatment (Fig. 7).
The cartoon illustrates the resistance mechanism of trastuzumab and the signal change by adavosertib in HER2-positive cancer. Left, shows PD-L1 upregulation contributes to trastuzumab resistance in HER2-positive cancer cells. Right, shows adavosertib can oversome the reastuzumab resistance through downregulation of PD-L1-CMTM6-BRCA1 signaling pathway. Adavosertib also reduces HER2-CDCP-1-SRC signals and immune related signals
Overall, our results demonstrated that upregulated PD-L1 on cancer cells contributed to trastuzumab resistance and could be reversed by WEE1 inhibition in HER2-positive cancer cells. The findings in this study support further clinical development of co-targeting DDR and HER2 for the treatment of patients with HER2-positive cancer.
References
Kim BW, Oh CM, Choi HY, Park JW, Cho H, Ki M. Incidence and overall survival of biliary tract cancers in South Korea from 2006 to 2015: using the National Health Information Database. Gut Liver. 2019;13:104–13.
Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64.
Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol. 2008;19:1523–9.
Merla A, Liu KG, Rajdev L. Targeted therapy in biliary tract cancers. Curr Treat Opt Oncol. 2015;16:48.
Vernieri C, Milano M, Brambilla M, Mennitto A, Maggi C, Cona MS, et al. Resistance mechanisms to anti-HER2 therapies in HER2-positive breast cancer: current knowledge, new research directions and therapeutic perspectives. Crit Rev Oncol Hematol. 2019;139:53–66.
Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–52.
Chaganty BKR, Qiu S, Gest A, Lu Y, Ivan C, Calin GA, et al. Trastuzumab upregulates PD-L1 as a potential mechanism of trastuzumab resistance through engagement of immune effector cells and stimulation of IFNgamma secretion. Cancer Lett. 2018;430:47–56.
Su S, Zhao J, Xing Y, Zhang X, Liu J, Ouyang Q, et al. Immune checkpoint inhibition overcomes ADCP-induced immunosuppression by macrophages. Cell. 2018;175(442–457):e423.
Tu X, Qin B, Zhang Y, Zhang C, Kahila M, Nowsheen S, et al. PD-L1 (B7-H1) Competes with the RNA exosome to regulate the DNA damage response and can be targeted to sensitize to radiation or chemotherapy. Mol Cell. 2019;74:1215–1226.
Arai H, Wada R, Ishino K, Kudo M, Uchida E, Naito Z. Expression of DNA damage response proteins in gastric cancer: comprehensive protein profiling and histological analysis. Int J Oncol. 2018;52:978–88.
Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–10.
Chen X, Low KH, Alexander A, Jiang Y, Karakas C, Hess KR, et al. Cyclin E overexpression sensitizes triple-negative breast cancer to Wee1 kinase inhibition. Clin Cancer Res. 2018;24:6594–610.
Lin X, Chen D, Zhang C, Zhang X, Li Z, Dong B, et al. Augmented antitumor activity by olaparib plus AZD1775 in gastric cancer through disrupting DNA damage repair pathways and DNA damage checkpoint. J Exp Clin Cancer Res. 2018;37:129.
Richer AL, Cala JM, O’Brien K, Carson VM, Inge LJ, Whitsett TG. WEE1 kinase inhibitor AZD1775 has preclinical efficacy in LKB1-deficient non-small cell lung cancer. Can Res. 2017;77:4663–72.
Jin MH, Nam AR, Park JE, Bang JH, Bang YJ, Oh DY. Therapeutic co-targeting of WEE1 and ATM downregulates PD-L1 expression in pancreatic cancer. Cancer Res Treat. 2020;52:149–66.
Nam AR, Kim JW, Cha Y, Ha H, Park JE, Bang JH, et al. Therapeutic implication of HER2 in advanced biliary tract cancer. Oncotarget. 2016;7:58007–21.
Jin MH, Nam AR, Park JE, Bang JH, Bang YJ, Oh DY. Resistance mechanism against trastuzumab in HER2-positive cancer cells and its negation by Src inhibition. Mol Cancer Ther. 2017;16:1145–54.
Zerdes I, Matikas A, Bergh J, Rassidakis GZ, Foukakis T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: biology and clinical correlations. Oncogene. 2018;37:4639–61.
Marti P, Stein C, Blumer T, Abraham Y, Dill MT, Pikiolek M, et al. YAP promotes proliferation, chemoresistance, and angiogenesis in human cholangiocarcinoma through TEAD transcription factors. Hepatology. 2015;62:1497–510.
Zhou Y, Huang T, Zhang J, Wong CC, Zhang B, Dong Y, et al. TEAD1/4 exerts oncogenic role and is negatively regulated by miR-4269 in gastric tumorigenesis. Oncogene. 2017;36:6518–30.
Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–5.
Alajati A, Guccini I, Pinton S, Garcia-Escudero R, Bernasocchi T, Sarti M, et al. Interaction of CDCP1 with HER2 enhances HER2-driven tumorigenesis and promotes trastuzumab resistance in breast cancer. Cell Rep. 2015;11:564–76.
You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010;20:402–9.
Dougan M, Dranoff G, Dougan SK. GM-CSF, IL-3, and IL-5 family of cytokines: regulators of inflammation. Immunity. 2019;50:796–811.
Hormigo A, Gu B, Karimi S, Riedel E, Panageas KS, Edgar MA, et al. YKL-40 and matrix metalloproteinase-9 as potential serum biomarkers for patients with high-grade gliomas. Clin Cancer Res. 2006;12:5698–704.
Vocka M, Langer D, Fryba V, Petrtyl J, Hanus T, Kalousova M, et al. Novel serum markers HSP60, CHI3L1, and IGFBP-2 in metastatic colorectal cancer. Oncol Lett. 2019;18:6284–92.
Oh DY, Bang YJ. HER2-targeted therapies—a role beyond breast cancer. Nat Rev Clin Oncol. 2020;17:33–48.
Mittal D, Vijayan D, Neijssen J, Kreijtz J, Habraken M, Van Eenennaam H, et al. Blockade of ErbB2 and PD-L1 using a bispecific antibody to improve targeted anti-ErbB2 therapy. Oncoimmunology. 2019;8:e1648171.
Loi S, Giobbie-Hurder A, Gombos A, Bachelot T, Hui R, Curigliano G, et al. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): a single-arm, multicentre, phase 1b–2 trial. Lancet Oncol. 2019;20:371–82.
Constantinidou A, Alifieris C, Trafalis DT. Targeting programmed cell death-1 (PD-1) and Ligand (PD-L1): a new era in cancer active immunotherapy. Pharmacol Ther. 2019;194:84–106.
Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27:409–16.
Wang Y, Wang H, Yao H, Li C, Fang JY, Xu J. Regulation of PD-L1: emerging routes for targeting tumor immune evasion. Front Pharmacol. 2018;9:536.
Zhang J, Dang F, Ren J, Wei W. Biochemical aspects of PD-L1 regulation in cancer immunotherapy. Trends Biochem Sci. 2018;43:1014–32.
Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine. 2018;35:233–43.
Zugazagoitia J, Liu Y, Toki M, McGuire J, Ahmed FS, Henick BS, et al. Quantitative assessment of CMTM6 in the tumor microenvironment and association with response to PD-1 pathway blockade in advanced-stage non-small cell lung cancer. J Thorac Oncol. 2019;14:2084–96.
Seifert AM, Reiche C, Heiduk M, Tannert A, Meinecke AC, Baier S, et al. Detection of pancreatic ductal adenocarcinoma with galectin-9 serum levels. Oncogene. 2020;39:3102–3113
Wang Y, Zhao E, Zhang Z, Zhao G, Cao H. Association between Tim3 and Gal9 expression and gastric cancer prognosis. Oncol Rep. 2018;40:2115–26.
Kubala MH, Punj V, Placencio-Hickok VR, Fang H, Fernandez GE, Sposto R, et al. Plasminogen activator inhibitor-1 promotes the recruitment and polarization of macrophages in cancer. Cell Rep. 2018;25(2177–2191):e2177.
Zhang Y, Lazarus J, Steele NG, Yan W, Lee HJ, Nwosu ZC, et al. Regulatory T-cell depletion alters the tumor microenvironment and accelerates pancreatic carcinogenesis. Cancer Discov. 2020;10:422–39.
Liu X, Zhou Z, Cheng Q, Wang H, Cao H, Xu Q, et al. Acceleration of pancreatic tumorigenesis under immunosuppressive microenvironment induced by Reg3g overexpression. Cell Death Dis. 2017;8:e3033.
Madhav A, Andres A, Duong F, Mishra R, Haldar S, Liu Z, et al. Antagonizing CD105 enhances radiation sensitivity in prostate cancer. Oncogene. 2018;37:4385–97.
Friedman J, Morisada M, Sun L, Moore EC, Padget M, Hodge JW, et al. Inhibition of WEE1 kinase and cell cycle checkpoint activation sensitizes head and neck cancers to natural killer cell therapies. J Immunother Cancer. 2018;6:59.
Sun L, Moore E, Berman R, Clavijo PE, Saleh A, Chen Z, et al. WEE1 kinase inhibition reverses G2/M cell cycle checkpoint activation to sensitize cancer cells to immunotherapy. Oncoimmunology. 2018;7:e1488359.
Souza-Fonseca-Guimaraes F, Cursons J, Huntington ND. The emergence of natural killer cells as a major target in cancer immunotherapy. Trends Immunol. 2019;40:142–58.
Wu SY, Fu T, Jiang YZ, Shao ZM. Natural killer cells in cancer biology and therapy. Mol Cancer. 2020;19:120.
Matheson CJ, Backos DS, Reigan P. Targeting WEE1 kinase in cancer. Trends Pharmacol Sci. 2016;37:872–81.
García-Parra J, Dalmases A, Morancho B, Arpí O, Menendez S, Sabbaghi M, et al. Poly (ADP-ribose) polymerase inhibition enhances trastuzumab antitumour activity in HER2 overexpressing breast cancer. Eur J Cancer. 2014;50:2725–34.
Wielgos ME, Zhang Z, Rajbhandari R, Cooper TS, Zeng L, Forero A, et al. Trastuzumab-resistant HER2(+) breast cancer cells retain sensitivity to poly (ADP-Ribose) polymerase (PARP) inhibition. Mol Cancer Ther. 2018;17:921–30.
Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, et al. PARP Inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res. 2017;23:3711–20.
Acknowledgements
This study was supported by Seoul National University Hospital research fund (Grant no. 03-2019-0220). This work has been supported by Institute of Smart Healthcare Innovative Medical Sciences, a Brain Korea 21 four program, Seoul National University
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
DY Oh is Consultant/Advisory Board members of AstraZeneca and Roche. No conflicts of interest were disclosed by the other authors.
Ethical approval
All institutional and national guidelines for the care and use of laboratory animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jin, MH., Nam, AR., Bang, JH. et al. WEE1 inhibition reverses trastuzumab resistance in HER2-positive cancers. Gastric Cancer 24, 1003–1020 (2021). https://doi.org/10.1007/s10120-021-01176-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-021-01176-7