Abstract
Background
The transcribed ultraconserved regions (T-UCRs) are a novel class of long non-coding RNAs and are involved in the development of several types of cancer. Although several different papers have described the oncogenic role of Uc.63+, there are no reports mentioning its importance in gastric cancer (GC) biology.
Methods
In this study, we evaluated Uc.63+ expression using clinical samples of GC by qRT-PCR, and also assessed the correlation between Uc.63+ expression and clinico-pathological factors.
Results
The upregulation of Uc.63+ was significantly correlated with advanced clinico-pathological features. Knockdown of Uc.63+ significantly repressed GC cell growth and migration, whereas overexpression of Uc.63+ conversely promoted those of GC cells. In situ hybridization of Uc.63+ revealed its preferential expression in poorly differentiated adenocarcinoma. We further conducted a microarray analysis using MKN-1 cells overexpressing Uc.63- and found that NF-κB signaling was significantly upregulated in accordance with Uc.63+ expression.
Conclusion
Our results suggest that Uc.63+ could be involved in GC progression by regulating GC cell growth and migration via NF-κB signaling
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) remains a deadly disease providing a great challenge in clinical management despite a steady decline in its incidence [1, 2]. Despite increased understanding of the genetic and epigenetic events involved in the pathogenesis of GC, the absence of solid methods for the diagnosis and treatment of GC remains one of the biggest difficulties in dealing with this disease [3, 4]. Additional in-depth analysis of the pathogenesis and biological features of GC is necessary to further enhance the elucidation of its molecular machinery, which could potentially lead to the improvement of current clinical care for GC.
Transcribed ultraconserved regions (T-UCRs) are genomic regions conserved across large evolutionary distances, which encode for non-coding RNAs that serve as regulators of gene expression. It has been reported that most of the T-UCRs found in normal human tissues are ubiquitous, tissue-specific, or both [5,6,7]. Growing evidence shows that T-UCRs are altered at the transcriptional level in human tumorigenesis, and the aberrant T-UCRs’ expression profiles can be used to differentiate human cancer types [8,9,10,11,12]. We have investigated the expression and molecular mechanism of T-UCRs in several cancers, and have revealed deregulation of a couple of T-UCRs in GC and prostate cancer (PC), some of which surely have contributed to the progression of the cancers [13,14,15]. Among them, Uc.63+ especially peaked our interest as it is a promising marker for the detection and stratification of resistance to docetaxel treatment of patients with PC [15]. In addition to our previous study, several different papers have also mentioned upregulation of Uc.63+ and the oncogenic role of Uc.63+ in breast and colorectal cancer [16, 17]; however, there have been no reports dealing with Uc.63+ in GC so far, to our knowledge. Moreover, neither of the published reports mentioned what signals or molecules interact with Uc.63+ expression in cancer biology.
The present study evaluated the expression and role of Uc.63+ in GC by analyzing its expression and distribution using qRT-PCR and in situ hybridization, and examining the relationship between Uc.63+ overexpression and clinico-pathological features. We also attempted to reveal the molecular mechanism that underlies the regulation of Uc.63+ expression by a microarray analysis and subsequent in silico analysis to help provide further supportive evidence of Uc.63+ expression as an indicator of GC with a more malignant phenotype.
Materials and methods
Patients and tissues
For qRT-PCR analysis, 40 pairs of GC and non-neoplastic gastric mucosa tissues were used. For in situ hybridization (ISH), archival formalin-fixed, paraffin-embedded tissues from 3 adenomas, and 10 intestinal-type and 10 diffuse-type GCs were used. All of the samples from primary tumors and adjacent non-neoplastic mucosa were collected from patients diagnosed as having GC who underwent surgery between 2005 and 2008 at Hiroshima University Hospital (Hiroshima, Japan). All samples were collected after obtaining appropriate informed consent from the patients. This study was approved by the Ethics Committee for Human Genome Research of Hiroshima University and conformed to the ethical guidelines of the Declaration of Helsinki. Tumor staging was determined according to the TNM classification system. Histological classification was carried out according to the Lauren classification [18].
qRT-PCR
Extraction of total RNA, synthesis of cDNA, and qRT-PCR were performed as described previously [19]. Briefly, total RNA was isolated from frozen samples or cancer cell lines using Isogen (Nippon Gene, Tokyo, Japan), and 1 μg of total RNA was converted to cDNA with a First Strand cDNA Synthesis Kit (Amersham Biosciences Corp., Piscataway, NJ, USA). The qPCR was performed with an SYBR Select Master Mix kit (Applied Biosystems, Austin, TX, USA). The primer sequences for the detection of all of the genes which we studied will be provided upon request. Real-time detection of the emission intensity of SYBR green bound to double-stranded DNA was performed with a CFX Connect Real-Time System (Bio-Rad Laboratories, Hercules, CA, USA). ACTB-specific PCR products, which were amplified from the same RNA samples, served as internal controls.
ISH
All of the procedures were performed as described previously [15]. Section 10-μm thick were rehydrated with xylene and 70–100% ethanol. Thereafter, sections were treated with 0.2-M sodium chloride and proteinase K. Slides were post-fixed, and sections were then demethylated with acetic anhydride and prehybridized. Hybridization was carried out in a humid chamber with 500 ng/mL of freshly prepared digoxigenin-labeled RNA probe of UC63. Sections were incubated for at least 48 h at 65 °C. The slides were washed, and incubation of the secondary anti-digoxigenin antibody (Roche Diagnostics, Indianapolis, IN, USA) was carried out at 4 °C overnight. The next day, sections were washed and developed using Nitro Blue Tetrazolium Chloride/5-Brom-4-Chlor-3-Indolyl-Phosphat (Roche Diagnostics).
Cell lines
Ten cell lines derived from human GC (MKN-1, MKN-7, MKN-45, MKN-74, HSC-57, TMK-1, KATO-III, HSC-39, HSC-44PE, and HSC-44As3) were used. HSC-39, HSC-57, HSC-44PE, and HSC-44As3 were established by Kazuyoshi Yanagihara [20, 21]. All cell lines were maintained in RPMI 1640 (05918; Nissui Pharmaceutical Co., Ltd., Tokyo) containing 10% fetal bovine serum (14-501F; BioWhittaker, Walkersville, MD, USA) in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. For low-oxygen condition, we cultured MKN-1 with 3% O2.
RNA interference (RNAi) and expression vector
Silencer® Select (Ambion, Austin, TX) against Uc.63+ was used for RNAi. Two independent oligonucleotides and negative control siRNA (Invitrogen, Carlsbad, CA, USA) were used. The sequence of siRNA#1 was 5′-AAAGAUGUUAACACUACCUga-3′, and that of siRNA#2 was 5′-UUUGGUGCUAAAUUUAUGCac-3′. The sequence of siRNA for Uc.160 + was described as previously [14]. In total, 1.0 × 106 cells of HSC-44As3 and MKN-1 were plated on a 10-cm culture dish 24 h before transfection. Transfection was performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Cells were used 48 h after transfection in each of the experiments and assays. For constitutive expression of Uc.63+, cDNA was PCR amplified and subcloned into pcDNA 3.1 (Invitrogen). The pcDNA-Uc.63+ expression vector was transfected into MKN-1 cells with FuGENE6 (Roche Diagnostics) according to the manufacturer’s instructions.
Cell growth assay and wound healing assay
To examine cell growth, an MTT assay was performed as described previously [22]. Cell growth was monitored after 1, 2, and 4 days.
Microarray analysis and bioinformatics
Total RNA from MKN-1 cells with forced expression of Uc.63+ and its control was isolated by lysing cells in Isogen lysis buffer (Nippon Gene) and further purified with RNeasy MinElute CleanUp Kit (QIAGEN). We performed microarray analyses using the Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA). The arrays were scanned with a GeneChip Scanner 3000 (Affymetrix), and the data were analyzed by Microarray Suite version 5.0 with Affymetrix default analysis settings and global scaling as the normalization method. Gene Set Enrichment Analysis (GSEA) (http://www.broadinstitute.org/gsea) was also applied for the analysis and validation of the gene expression profiles.
Western blotting
Western blotting was performed as described previously [23]. Immunocomplexes were visualized with an ECL Western blot detection system (RPN2109; Amersham Biosciences, Buckinghamshire, UK). β-Actin (A5316; Sigma-Aldrich) was also stained as a loading control.
Immunohistochemistry (IHC)
Immunostaining was performed as described previously [23]. Anti‐NF‐κB p65 antibody was purchased from Cell Signaling Technology (D14E12, 1:500; Danvers, MA, USA).
Kaplan–Meier analysis
Kaplan–Meier analysis was performed for RELA using the KMplot software from a database of public microarray data sets (http://kmplot.com/analysis). The results from 876 GC patients were collected from this database. Kaplan–Meier analysis was performed for Uc.63 (chromosome 2. 61727035-61727312) using TCGA software from a database of public RNA-sequencing data sets (http://kmplot.com/analysis). The results from 253 GC patients were collected from this database. To analyze the prognostic value of the probe, the samples were split into two groups according to the cutoff value generated by the software. Hazard ratios and p values (log-rank p) are shown for each survival analysis.
Statistical methods
Statistical differences were evaluated using a two-tailed Student’s t test or Mann–Whitney U test. The correlation between expression levels of Uc.63+ and clinico-pathological characteristics was analyzed with Fisher’s exact test. The results are expressed as the mean ± SD of triplicate measurements. A p value of less than 0.05 was considered statistically significant.
Results
Uc.63+ expression in GC and its clinico-pathological significance
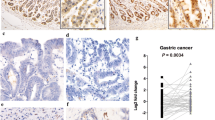
We compared the expression of Uc.63+ in 40 GC tissues and corresponding normal stomach tissues by qRT-PCR. As shown in Fig. 1a, most of the GC tissues showed higher Uc.63+ expression than those of normal stomach. When we regarded a GC tissue/normal stomach tissue ratio > 2.0 as upregulation, 21 of 40 GC cases showed upregulation of Uc.63+ (Fig. 1b). We also evaluated the correlation between Uc.63+ expression and clinico-pathological characteristics in the 40 GC cases, and found that higher Uc.63+ expression was significantly correlated with advanced T grade, N grade, stage, and a tendency to show diffuse-type histology (Table 1). We then looked into the correlation between Uc.63 expression and GC patient’s survival using TCGA data base, and found that there was no significant correlation between them (Figure S1). To further validate the data obtained from qRT-PCR-based studies, we performed ISH using GC tissue sections with 10 intestinal-type and 10 diffuse-type histologies, respectively. The areas displaying non-neoplastic mucosa, including fundic glands and intestinal metaplasia, and adenomas did not show any obvious signals (Fig. 1c). In the cancerous areas, we could detect a robust signal of Uc.63+ in 9 of 10 diffuse-type GC cases, whereas almost no signals were detectable in all 10 of the intestinal-type GC cases (Fig. 1c), which was consistent with the results obtained with qRT-PCR. Concerning the distribution of Uc.63+ expression in the tissue section, most of the cancer cells displaying diffuse-type histology in Uc63 + positive GC cases showed detectable signals of Uc.63+ regardless of localization such as intramucosal region and invasive front. One of the points that piqued our interest is that the cancer cells displaying intestinal-type histology even in Uc63 + positive GC cases that are mainly composed of diffuse-type histology did not showed detectable signals of Uc.63+, which further support the results obtained with qRT-PCR. These data indicated that Uc.63+ contributes to the progression of GC and tends to express in GC tissues with diffuse-type histology.
Uc.63+ expression in gastric cancer (GC) and its clinico-pathological significance. a The qRT-PCR results for Uc.63+ in 40 GC tissues and corresponding normal mucosa. b T/N ratio of UC63 expression level between GC tissue (T) and corresponding non-neoplastic mucosa (N) in 40 GC cases. A T/N ratio greater than 2 was considered to represent overexpression. c In situ hybridization of Uc.63+ in non-neoplastic mucosa, adenoma and GC tissue. Left upper panel: fundic gland. Left middle panel: intestinal metaplasia. Left lower panel: gastric adenoma. Middle and right upper panels: low and high magnification of intestinal-type GC. Middle and right middle panels: low and high magnification of diffuse-type GC case (solid type). Middle and right lower panels: low and high magnification of diffuse-type GC cases (mucinous type). Scale bars indicate (left) 100 µm, (middle) 100 µm, and (right) 10 µm
Preference of Uc.63+ expression in GC cell lines and correlation with cell growth
To further verify the aforementioned findings, we investigated the functional role of Uc.63+ in GC. We first examined Uc.63+ expression in 5 non-neoplastic gastric mucosa and 10 GC cell lines. All of the GC cell lines showed high Uc.63+ expression compared to that in normal tissue. The cell lines derived from diffuse-type GC especially showed higher Uc.63+ expression than that in GC cells from the other types GC (Fig. 2a), which further supported the preference of Uc.63+ expression in diffuse-type GC. Although one previous study reported that Uc.63+ was induced by hypoxia in colorectal cancer, there have been no reports concerning the regulatory mechanism of Uc.63+ expression in cancer. We cultured MKN-1 cell in the incubator with 3% O2, and found that both HIF1A and Uc.63+ were significantly upregulated (Figure S2A). To elucidate the biological function of Uc.63+ in GC, we transfected a vector containing the whole sequence of Uc.63+ into MKN-1 cells that possessed the lowest level of Uc.63+ expression among the 10 GC cell lines and established two different clones of MKN-1 with the forced expression of Uc.63+ . Transfection of the vector successfully induced Uc.63+ overexpression in both of the clones (Fig. 2b). We then performed an MTT assay and found that MKN-1 cells transfected with the Uc.63+ expression vector showed significantly enhanced cell proliferation (Fig. 2c). We previously reported that down regulation of Uc.160 + is involved in GC progression [14]. When we inhibited Uc.160 + by a specific siRNA in concert with transfection of Uc.63+ expression vector, cell proliferation was significantly enhanced compared to that of only Uc.63+ overexpression (Figure S2B). To further determine the importance of Uc.63+ in GC cell proliferation, we investigated the effects of Uc.63+ knockdown by siRNAs that specifically target Uc.63+. We confirmed a significantly lower Uc.63+ expression in HSC-44As3 cells with two different siRNAs than that in the negative control (Fig. 2d). In the MTT assay, we found that Uc.63+ knockdown inversely reduced cell proliferation (Fig. 2e). These results support the likely probability of Uc.63+ to have an oncogenic role in GC.
Preference of Uc.63+ expression in gastric cancer (GC) cell lines and correlation with cell growth. a The qRT-PCR results for Uc.63+ in 5 non-neoplastic gastric tissues and 10 GC cell lines. b The qRT-PCR for Uc.63+ in MKN-1 cells transfected with Uc.63+ expression vector or empty vector. The results are expressed as the mean ± SD of triplicate measurements. c MTT assay in MKN-1 cells transfected with Uc.63+ expression vector or empty vector. Cell growth was assessed at 1, 2, and 4 days after seeding on 96-well plates. Bars and error bars are the mean ± SD, respectively, of triplicate measurements. d The qRT-PCR results for Uc.63+ in HSC-44As3 cells transfected with negative control or two different siRNAs. The results are expressed as the mean ± SD of triplicate measurements. e MTT assay in the negative control and two different siRNAs. Cell growth was assessed at 1, 2, and 4 days after seeding on 96-well plates. Bars and error bars are the mean ± SD, respectively, of triplicate measurements
Search for genes and molecular pathways influenced by Uc.63+ overexpression using microarray and in silico analysis
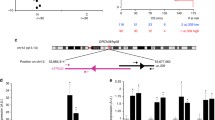
To further scrutinize the molecular basis of the GC phenotypes driven by Uc.63+ overexpression, we performed a microarray analysis using 2 clones of MKN-1 with Uc.63+ overexpression and its controls. The expression profile was mainly validated in GSEA. We identified 368 upregulated genes (p < 0.01 and > twofold change) and 194 downregulated genes (p < 0.01 and < 0.5-fold change). The list of 20 genes that showed specific upregulation are summarized in Table 2. Notably, many chemokine-related genes, such as IL6 and CCL2, were upregulated in accordance with Uc.63+ overexpression which was further supported by Gene Ontology (GO) analysis that indicated a significant alteration of several chemokine-related GO terms including “cytokine activity” and “Interleukin-1 receptor binding”. To further clarify the phenotype of MKN-1 with Uc.63+ overexpression, we performed GSEA analysis especially focusing on the above-mentioned 368 upregulated genes that showed specific up-regulation along with Uc.63+ overexpression, and determined a significant enrichment of gene sets for “HINATA_NFKB_IMMU_INF” in MKN-1 with Uc.63+ overexpression (Table 3 and Fig. 3a). We chose the top 8 upregulated genes that were involved in chemokine activity from the list of upregulated genes (Fig. 3b), and their upregulation was further validated by qRT-PCR (Fig. 3c). We then examined the effect of Uc.63+ on the expression of p65, which is one of the important subunits in the NF-κB complex. As shown in Fig. 3d, expression of p65 was obviously induced by Uc.63+ overexpression. To further confirm the expression of p65, we performed IHC and detected robust expression in the nucleus and the specific siRNA for RELA, which is the coding gene of p65, significantly silenced p65 expression (Figure S3A). To further determine the importance of p65 expression in GC cell proliferation, we performed MTT assay using above-mentioned 2 MKN-1 cells showed in Figure S3A and found that knockdown of p65 significantly inhibited cell growth activity. We found that Uc.63+ knockdown inversely reduced cell proliferation (Fig. 2e). We also examined the relation between expression of RELA and the prognosis of the patients with GC using a public on-line database. We found that GC cases with higher expression of RELA showed significantly worse prognosis than those with lower expression of RELA (Fig. 3e), which further supported that the Uc.63+/NF-κB axis contributes to GC progression and can lead to poor clinical outcome of GC patients.
Search for the genes and molecular pathways influenced by Uc.63+ overexpression using microarray and in silico analysis. a GSEA of MKN-1 with Uc.63+ overexpression versus its control, highlighting the enrichment of NF-κB-related genes. b A heat map showing chemokine-related genes in MKN-1 with Uc.63+ overexpression and its control. c The qRT-PCR results for chemokine-related genes in MKN-1 with Uc.63+ overexpression and its control. d Western blot analysis of p65 in MKN-1 with Uc.63+ overexpression and its control. β-Actin was used as a loading control. e Kaplan–Meier analysis of the probability of overall survival in GC patients according to RELA expression. Hazard ratios (HRs) and p values (log-rank test) are shown for each survival analysis
Discussion
The results of the present study showed the clinico-pathological significance of Uc.63+ expression in GC and provided evidence that GC cases with Uc.63+ overexpression are more likely to be related to advanced cancer and that Uc.63+ overexpression is preferentially seen in diffuse-type GC. To our knowledge, only two studies have mentioned the clinical significance of Uc.63+ expression: Marini et al. revealed that higher Uc.63+ expression was significantly correlated with poor clinical outcome of breast cancer patients [16], and we previously identified that high levels of Uc.63+ expression were frequently detected in patients with castration-resistant PC and docetaxel-resistant PC [15], both of which demonstrated that Uc.63+ may play an oncogenic role in cancer tissues and are consistent with our present data in GC. In the present study, we found that there was no significant correlation between the correlation between Uc.63 expression and GC patient’s survival using TCGA database. However, one issue is that the expression of Uc.63 was determined with whole transcripts from chromosome 2. 61727035–61727312, which means that expression of both Uc.63+ and Uc.63+ A would be evaluated in the study. To precisely evaluate the direct correlation between Uc.63+ and GC patients’ survival, we could examine Uc.63+ expression using GC samples with the information of patient’s prognosis. Although our results indicated that high Uc.63+ expression was preferentially seen in diffuse-type GC tissues and cells, we could not obtain any clues implying the molecular basis that underpin the diffuse-type morphology of GC from either the microarray or the in silico analysis. In breast cancer, higher Uc.63+ expression was significantly associated with poor prognosis only in luminal type A breast cancer cases [16], which also indicates that Uc.63+ could play a functional role only in some specific histological types of cancers. Further in-depth studies are still needed to characterize the mechanism underlying the correlation between Uc.63+ expression and pathological characteristics of the cancers, which may also reveal the fundamental mechanisms commonly involved in the progression of several types of cancers.
Another important finding of this study is that Uc.63+ overexpression significantly induced activation of NF-κB signaling. In terms of the regulatory machinery of Uc.63+ expression, evidence indicated that Uc.63+ expression was regulated by hypoxia [17], and the plausible promoter region of Uc.63+ was thought to have a binding site of hypoxia-inducible factor [16]. As for the downstream targets of Uc.63+, we previously revealed that Uc.63+ regulated the expression of MMP2 through its interaction with miR-130b regulation [15]. As far as we know, there is little evidence of Uc.63+ being implicated in the regulation of some specific genes and/or signalings; this is the first study to report the direct effect of T-UCRs on NF-κB signaling and its downstream target p65. However, one of the limitations of this study is that we could not clarify whether Uc.63+ directly affects the expression of the components of the NF-κB signaling pathway. Several lines of study have provided evidence for an important role of the interaction between T-UCRs and miRNA in cancer biology [24,25,26], and we have actually identified the tight interaction between Uc.63+ and miR-130b. By reviewing TargetScanHuman (http://www.targetscan.org/vert_72/) to find candidates of target genes of miR-130b, several NF-κB-related genes, such as TNFRSF11A [27] and NKAP [28], were listed (data not shown); further validation of this list may lead to the identification of solid evidence concerning the involvement of Uc.63+ in the regulation of NF-κB signaling. The other possible explanation is that there could probably be many miRNAs that are potentially regulated by Uc.63+, which may also help to inform how Uc.63+ is involved in the regulation of NF-κB signaling. Taken together, these findings call for additional studies on the interaction between Uc.63+ and miRNAs to determine all the aspects of the molecular mechanism in cancer biology.
In closing, the present study verified the clinico-pathological significance of Uc.63+ overexpression in GC and also proved the oncogenic role of Uc.63+ through the regulation of NF-κB signaling. Although the details of the interaction between Uc.63+ and NF-κB signaling in GC remain unclear, these data highlight its pivotal role in the progression of GC.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Venerito M, Vasapolli R, Rokkas T, Malfertheiner P. Gastric cancer: epidemiology, prevention, and therapy. Helicobacter. 2018;23(Suppl 1):e12518.
Shimizu D, Kanda M, Kodera Y. Review of recent molecular landscape knowledge of gastric cancer. Histol Histopathol. 2018;33:11–26.
Ajani JA, Lee J, Sano T, Janjigian YY, Fan D, Song S. Gastric adenocarcinoma. Nat Rev Dis Prim. 2017;3:17036.
Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA Biol. 2013;10:1771–7.
Scaruffi P, Stigliani S, Coco S, Valdora F, De Vecchi C, Bonassi S, et al. Transcribed-ultra conserved region expression profiling from low-input total RNA. BMC Genom. 2010;11:149.
Jiang BC, Yang T, He LN, Tao YX, Gao YJ. Altered T-UCRs expression profile in the spinal cord of mice with neuropathic pain. Transl Perioper Pain Med. 2016;1:1–10.
Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29.
Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–401.
Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, Kogure T, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:786–91.
Jiang J, Azevedo-Pouly AC, Redis RS, Lee EJ, Gusev Y, Allard D, et al. Globally increased ultraconserved noncoding RNA expression in pancreatic adenocarcinoma. Oncotarget. 2016;7:53165–77.
Galasso M, Dama P, Previati M, Sandhu S, Palatini J, Coppola V, et al. A large scale expression study associates uc.283-plus lncRNA with pluripotent stem cells and human glioma. Genome Med. 2014;6:76.
Goto K, Ishikawa S, Honma R, Tanimoto K, Sakamoto N, Sentani K, et al. The transcribed-ultraconserved regions in prostate and gastric cancer: DNA hypermethylation and microRNA-associated regulation. Oncogene. 2016;35:3598–606.
Honma R, Goto K, Sakamoto N, Sekino Y, Sentani K, Oue N, et al. Expression and function of U.c160 +, a transcribed ultraconserved region, in gastric cancer. Gastric Cancer. 2017;20:960–9.
Sekino Y, Sakamoto N, Goto K, Honma R, Shigematsu Y, Sentani K, et al. Transcribed ultraconserved region U.c63+ promotes resistance to docetaxel through regulation of androgen receptor signaling in prostate cancer. Oncotarget. 2017;8:94259–70.
Marini A, Lena AM, Panatta E, Ivan C, Han L, Liang H, et al. Ultraconserved long non-coding RNA u.c63 in breast cancer. Oncotarget. 2017;8:35669–80.
Ferdin J, Nishida N, Wu X, Nicoloso MS, Shah MY, Devlin C, et al. HINCUTs in cancer: hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ. 2013;20:1675–87.
Lauren P. the two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49.
Sakamoto N, Naito Y, Oue N, Sentani K, Uraoka N, Zarni Oo H, et al. MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Sci. 2014;105:236–43.
Yanagihara K, Tanaka H, Takigahira M, Ino Y, Yamaguchi Y, Toge T, et al. Establishment of two cell lines from human gastric scirrhous carcinoma that possess the potential to metastasize spontaneously in nude mice. Cancer Sci. 2004;95:575–82.
Yanagihara K, Takigahira M, Tanaka H, Komatsu T, Fukumoto H, Koizumi F, et al. Development and biological analysis of peritoneal metastasis mouse models for human scirrhous stomach cancer. Cancer Sci. 2005;96:323–32.
Sakamoto N, Oue N, Noguchi T, Sentani K, Anami K, Sanada Y, et al. Serial analysis of gene expression of esophageal squamous cell carcinoma: ADAMTS16 is upregulated in esophageal squamous cell carcinoma. Cancer Sci. 2010;101:1038–44.
Sakamoto N, Oue N, Sentani K, Anami K, Uraoka N, Naito Y, et al. Liver–intestine cadherin induction by epidermal growth factor receptor is associated with intestinal differentiation of gastric cancer. Cancer Sci. 2012;103:1744–50.
Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, et al. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology. 2018;154:599–611.
Wang JY, Cui YH, Xiao L, Chung HK, Zhang Y, Rao JN, et al. Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol. 2018;38:e00010–8.
Terreri S, Durso M, Colonna V, Romanelli A, Terracciano D, Ferro M, et al. New cross-talk layer between ultraconserved non-coding RNAs, microRNAs and polycomb protein YY1 in bladder cancer. Genes. 2016;7:E127.
Santini D, Schiavon G, Vincenzi B, Gaeta L, Pantano F, Russo A, et al. Receptor activator of NF-kB (RANK) expression in primary tumors associates with bone metastasis occurrence in breast cancer patients. PLoS One. 2011;6:e19234.
Cuesta S, Kireev R, Forman K, Garcia C, Escames G, Ariznavarreta C, et al. Melatonin improves inflammation processes in liver of senescence-accelerated prone male mice (SAMP8). Exp Gerontol. 2010;45:950–6.
Acknowledgements
We thank Mr. Shinichi Norimura for his technical assistance. This work was carried out with the kind cooperation of the Research Center for Molecular Medicine of the Faculty of Medicine of Hiroshima University. We also thank the Analysis Center of Life Science of Hiroshima University for the use of their facilities.
Funding
This work was supported by the Grants-in-Aid for Scientific Research (JP15H04713, JP16K08691, JP16H06999) and Challenging Exploratory Research (26670175, JP16K15247) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human rights statement
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions.
Informed consent
Informed consent or substitute for it was obtained from all patients for being included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10120_2020_1070_MOESM1_ESM.tif
Supplementary material 1 Kaplan–Meier analysis of the probability of overall survival in GC patients according to Uc63 expression. p-values (log rank test) are shown for each survival analysis. (TIFF 537 kb)
10120_2020_1070_MOESM2_ESM.tif
Supplementary material 2 (A) The qRT-PCR results for HIF1A and Uc.63+ cultured in the incubators with (left: blue) 20% O2 and (right: red) 3% O2, respectively. The results are expressed as the mean ± S.D. of triplicate measurements. (B) MTT assay in MKN-1 cells transfected with Uc.63+ expression vector or empty vector in concert with the specific siRNA for Uc.160+. Cell growth was assessed at 1, 2 and 4 days after seeding on 96-well plates. Bars and error bars are the mean ± S.D., respectively, of triplicate measurements. (TIFF 640 kb)
10120_2020_1070_MOESM3_ESM.tif
Supplementary material 3 (A) Immunohistochemical staining of p65 using MKN-1 with (left) Uc.63+ overexpression and (right) Uc.63+ overexpression with siRNA treatment for RELA. Scale bars indicate 10um. (B) MTT assay using MKN-1 with Uc.63 overexpression in the negative control and the siRNA for RELA. Cell growth was assessed at 1, 2 and 4 days after seeding on 96-well plates. Bars and error bars are the mean ± S.D., respectively, of triplicate measurements. (TIFF 4418 kb)
Rights and permissions
About this article
Cite this article
Sakamoto, N., Sekino, Y., Fukada, K. et al. Uc.63+ contributes to gastric cancer progression through regulation of NF-kB signaling. Gastric Cancer 23, 863–873 (2020). https://doi.org/10.1007/s10120-020-01070-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-020-01070-8