Abstract
Background
Exploratory biomarker analysis was conducted to identify factors related to the outcomes of patients with stage II/III gastric cancer using data from the Adjuvant Chemotherapy Trial of S-1 for Gastric Cancer, which was a randomized controlled study comparing the administration of an orally active combination of tegafur, gimeracil, and oteracil with surgery alone.
Methods
Formalin-fixed paraffin-embedded surgical specimens from 829 patients were retrospectively examined, and 63 genes were analyzed by quantitative real-time RT-PCR after TaqMan assay-based pre-amplification. Gene expression was normalized to the geometric mean of GAPDH, ACTB, and RPLP0 as reference genes, and categorized into low and high values based on the median. The impact of gene expression on survival was analyzed using 5-year survival data. The Benjamini and Hochberg procedure was used to control the false discovery rate.
Results
IGF1R and AREG were most strongly correlated with overall survival, which was significantly worse in high IGF1R patients than low IGF1R patients, but better in high AREG patients than low AREG patients. The hazard ratio for death in the analysis of overall survival (S-1 vs. surgery alone) was reduced in the high IGF1R group compared with the low IGF1R group and in the low AREG group compared with the high AREG group. There were no significant interaction effects.
Conclusion
IGF1R gene expression was associated with poor outcomes after curative resection of stage II/III gastric cancer, whereas AREG gene expression was associated with good outcomes. No significant interaction effect on survival was evident between S-1 treatment and gene expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite a decreasing trend in Japan, gastric cancer remains the second most common cause of cancer-related death worldwide. Adequate surgery is the only treatment known to offer a cure, with adjuvant therapy improving overall survival (OS). The Adjuvant Chemotherapy Trial of S-1 for Gastric Cancer (ACTS-GC), which was a prospective randomized phase III trial, demonstrated that surgery plus treatment with an orally active combination of tegafur, gimeracil, and oteracil at a molar ratio of 1:0.4:1 (TS-1; Taiho Pharmaceutical, Tokyo, Japan) was more effective than surgery alone in Japanese patients with stage II/III gastric cancer [1–3]. The 5-year OS rate was 71.7 % in the S-1 group versus 61.1 % in the surgery-alone group. However, the 5-year overall survival (OS) rate in patients with stage IIIB disease was 50.2 % in the S-1 group in a subset analysis, suggesting room for improvement. This finding highlighted the need to identify the factors influencing relapse to develop more effective treatments for high-risk gastric cancer patients.
We have already published two papers on ACTS-GC biomarker studies. Terashima et al. tested HER2 and EGFR for their potential as markers by performing an immunohistochemical assay, and reported that EGFR is a poor prognostic marker, and not a predictive marker [4]. Sasako et al. tested genes involved in pyrimidine metabolism (TS, DPD, OPRT, TP) for their potential as predictive markers by performing an RT-PCR assay, and reported that TS and DPD mRNAs are better predictive markers [5]. The methods used in these two articles together constitute the so-called candidate approach. In the present study, we expanded the number of genes up to 63, compared with the aforementioned candidate approach that used only a few genes, to investigate a prognostic or predictive marker for S-1 therapy. We have included genes encoding key molecules such as those involved in growth factor signaling pathways, apoptotic signaling pathways, and DNA repair mechanisms, as well as 5-FU-related genes. This method is based on previous reports, which showed that the molecules involved in growth factor signaling pathways, apoptotic signaling pathways, and DNA repair mechanisms served as prognostic factors and significant predictive markers in the development of the fluorinated pyrimidine-based anticancer agent against stomach cancer [6, 7]. Thus, we could perform hypothesis-driven testing of the panel of 63 genes selected on the basis of their biological functions and relationships reported in the literature. Furthermore, in previous reports published by Sasako, a real-time RT-PCR technique without pre-amplification was used for mRNA detection. In the present study, we used a highly sensitive detection procedure involving multiplex pre-amplification of 14 cycles before real-time PCR detection with TaqMan Array Cards on FFPE samples. This procedure enabled us to detect low gene expression levels more precisely than did the previous procedure, where lower gene expression levels were not detected. Thus, we retrospectively evaluated whether they were predictive markers for the response to S-1 and/or prognostic markers for patients enrolled in the ACTS-GC.
Materials and methods
Study population and design
Tumor tissue was collected from patients enrolled in the ACTS-GC, the inclusion criteria and treatment protocol of which have been described previously [4, 5]. After the completion of the first interim analysis of the ACTS-GC, this biomarker study was designed retrospectively to determine any predictive value for the benefit of S-1 treatment or for prognosis. The protocol used for the current biomarker study was approved by the ethics committee of the Japanese Gastric Cancer Association and the institutional review board of each participating hospital, and complied with the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) guidelines [8].
Reverse-transcription PCR
Hematoxylin and eosin-stained slides from formalin-fixed, paraffin-embedded (FFPE) specimens were reviewed by a pathologist to estimate the tumor load. Sections (10 μm thick) were then stained with nuclear fast red (Sigma-Aldrich, St. Louis, MO, USA) for manual microdissection. Tumor tissue was selected at a magnification of 5× to 10× and dissected using a scalpel, as described previously [9]. RNA was isolated from tumor tissue and cDNA was prepared as described previously [9], with a slight modification in the extraction step, which used RNeasy Mini Elute spin-columns (Qiagen, Chatsworth, GA, USA). The expression levels of 63 genes were determined using TaqMan real-time PCR (TaqMan array card; Life Technologies, Foster City, CA, USA) after TaqMan assay-based pre-amplification. Briefly, cDNA (2.5 µl) was pre-amplified using TaqMan PreAmp Master Mix (2×) (Life Technologies) and a pool of TaqMan Gene Expression Assays (0.2×) in a 10-µl polymerase chain reaction (PCR). The pre-amplification cycling conditions were as follows: 95 °C for 10 min, followed by 14 cycles of 95 °C for 15 s, and 60 °C for 4 min. An amplified cDNA sample was diluted 20 times in TE buffer. Amplified cDNA (25 µl) was added to 25 µl RNase-free water and 50 µl 2× TaqMan Gene Expression Master Mix (Life Technologies). The mixture was then transferred to a loading port for the TaqMan low-density array (LDA). The LDA was centrifuged twice, sealed, and PCR amplification was performed using the Applied Biosystems Prism 7900HT Sequence Detection System (Life Technologies) under the following thermal cycling conditions: 50 °C for 2 min and 94.5 °C for 10 min, followed by 40 cycles of 97 °C for 30 s and 59.7 °C for 1 min. The LDA included ACTB, GAPDH, and RPLP0 as references based on their proven role as housekeeping genes [10, 11]. The assay IDs used in the LDA are shown in supplemental Table S1. The cycle threshold (C t) value, which is inversely proportional to the amount of cDNA, was calculated. The gene expression (relative mRNA) levels were expressed as the ratios (the differences between the C t values) between the gene of interest and the geometric mean of the reference genes, which provided a baseline measurement for the amount of mRNA isolated from a specimen. The expression levels of each gene were categorized as low or high based on the 50th percentile (median). The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines checklist used is shown in supplemental Table S2.
Data processing and statistical analysis
To determine the stability of the reference genes, the geNORM algorithm (MS-Excel add-on-macro program) was used, as described previously by Vandesompele et al. [12]. The program calculates the gene stability measure M by determining the average pairwise variation between a particular reference gene and all other control genes. Using genes with M values lower than 1.5, a normalization factor was calculated based on the geometric mean of the expression levels of the selected genes. To control the quality, target genes with data obtained from more than 60 % of the samples were employed, and the rest were excluded from further analysis.
The categorical data were analyzed using the chi-square test. Either the Wilcoxon or the Kruskal–Wallis test was used to assess correlations between groups. Survival curves were estimated using the Kaplan–Meier product-limit method, and the statistical significance of differences between survival curves was assessed using the log-rank test. Univariate and multivariate survival analyses were performed using a Cox proportional hazards model. Results were considered statistically significant at P < 0.05. All statistical analyses used the SAS software package version 9.1, the JMP software version 8.01 (SAS Institute, Cary, NC, USA) and MS-Excel (add-on-macro program; Microsoft, Redmond, WA, USA). The Benjamini and Hochberg false discovery rate (FDR)-controlling procedure was employed for multiple comparisons. Correlations of gene expression and prognosis were considered statistically significant at FDR P < 0.10.
Validation of the prognostic capability of selected genes in an independent data set
Publicly available Illumina-DASL gene expression and clinical data (RFS data only; OS data were not available) of 432 samples from gastric cancer patients in Asia were downloaded via the Gene Expression Omnibus (GEO) database accession number GSE26253 [13]. Raw data of the GSE26253 data set were loaded onto GeneSpring GX version 12.6 (Agilent Technologies, Santa Clara, CA, USA). Gene expression data were normalized by two strategies: “per chip normalization” and “per gene normalization.” For “per chip normalization,” all expression data on a chip were normalized to the 75th percentile of all values on that chip. For “per gene normalization,” the data for a given gene were normalized to the median expression level of that gene across all samples. The detection P value was utilized for subsequent data quality control (QC) procedures. According to gene expression levels summarized from QC-passed probes (detection P < 0.05, >50 % of samples), 297 stage II or III gastric cancers from all 432 samples were categorized into two groups (i.e., “High” or “Low,” compared to the median) and were subjected to survival analysis as previously mentioned.
Results
Patient characteristics
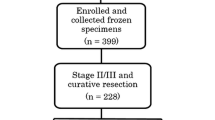
Archived FFPE specimens obtained by surgical resection were available for 829 (78.3 %) of the 1059 patients who were enrolled in the ACTS-GC at 65 centers and constituted the biomarker study population. A summary of the patient demographic data and tumor characteristics was published elsewhere (Supplemental Table 3) [4, 5]. The median patient age was 62 years (range, 27–80 years). There was no significant difference between the population used in the current biomarker study and the total population of the ACTS-GC, as previously reported [2].
Gene expression
The gene expression stability measures for the reference genes were calculated as 0.916, 0.931, and 0.923 for GAPDH, ACTB, and RPLP0, respectively. The M values were lower than 1.5 for all three genes, indicating that they could be utilized to normalize the target genes. Four genes (CDKNA2, EGF, IGF2, SEMA3B) were excluded from further analysis because their expression levels were below the detection limit in less than 60 % of the samples (51.5 %, 23.6 %, 44.5 %, and 5.5 %, respectively). Thus, 59 of the 63 genes subjected to LDA passed the quality control criteria. The median success rate for the 59 genes measured was 98.6 % (range, 61.2–100 %).
Overall correlation of gene expression and OS or relapse-free survival
Table 1 shows that among the 56 screened genes (excluding the three reference genes), IGF1R and AREG were most strongly correlated with OS (FDR, 0.0048 and 0.018, respectively). Kaplan–Meier plots of OS for all patients according to IGF1R and AREG expression levels are shown in Fig. 1. OS was significantly worse in high IGF1R patients than in low IGF1R patients, but better in high AREG patients than in low AREG patients. IGF1R was most strongly correlated with relapse-free survival (RFS; FDR, 0.007; Supplemental Table S4). Kaplan–Meier plots of the RFS of all patients according to IGF1R expression levels are shown in Supplemental Figure S1. RFS was significantly worse in high IGF1R patients than in low IGF1R patients.
Correlation of gene expression with OS or RFS in each treatment arm
Table 2 shows that among the 56 screened genes, only IGF1R was correlated with OS for patients who received surgery alone (FDR, 0.01). Kaplan–Meier plots of OS in the surgery-only arm are shown in Supplemental Figure S2A. OS was significantly worse in high IGF1R patients than in low IGF1R patients. No statistically significant correlations were detected between gene expression and OS in the S-1 arm. Supplemental Table S5 shows that IGF1R was correlated with RFS for patients who received surgery alone (FDR, 0.020). Kaplan–Meier plots of RFS in the surgery-alone arm are shown in Supplemental Figure S2B. RFS was significantly worse in high IGF1R patients than in low IGF1R patients. No statistically significant correlations were observed between gene expression and RFS in the S-1 arm.
Predictive value of biomarker analysis
Kaplan–Meier plots of OS for S-1 treatment versus surgery alone according to IGF1R and AREG expression levels are shown in Fig. 2a–d. The hazard ratio (HR) for death in the analysis of OS (S-1 vs. surgery alone) was lower in the high IGF1R group (HR, 0.55; 95 % CI, 0.40–0.76) than in the low IGF1R group (HR, 0.72; 95 % CI, 0.49–1.06). Similarly, the HR for death in the analysis of OS (S-1 vs. surgery alone) was much smaller in the low AREG group (HR, 0.57; 95 % CI, 0.41–0.79) than in the high AREG group (HR, 0.74; 95 % CI, 0.51–1.08). The prognostic relevance of IGF1R and AREG was assessed using a multivariate proportional hazards model adjusted for the following established clinical prognostic factors: treatment arm, gender, age, cancer stage, and histological type (Table 3). Although treatment arm and cancer stage were strong prognostic factors, IGF1R and AREG status were also independent prognostic factors. No statistically significant interactions were observed between IGF1R or AREG expression and S-1 treatment (Fig. 3).
Correlations among gene expressions and clinicopathological parameters
There was no statistically significant correlation between the mRNA expression levels of IGF1R and AREG (Spearman’s rank correlation coefficient: r = 0.035), and AREG and EGFR (r = 0.16). Any statistically significant relationship was not observed between clinicopathological parameters such as T, N, grade of differentiation, histological subtype, tumor size, and IGF1R and AREG gene expressions.
Validation of prognostic capability of selected genes in an independent data set
QC-passed microarray data of an independent study cohort validated the prognostic capability of 44 genes of the 56 genes screened by LDA (Supplemental Table S6).
Twelve genes including AREG did not pass the QC procedure. Expression levels of SPARC, EZH2, IGF1R, and E2F1 were strongly correlated with RFS (HR, 1.81, 0.63, 1.49, and 0.67; FDR, 0.06, 0.26, 0.34, and 0.34, respectively). Kaplan–Meier plots of RFS for all patients according to SPARC, EZH2, IGF1R, and E2F1 expression levels are shown in Supplemental Figure S3. RFS was worse in patients with high SPARC or IGF1R than in patients with low SPARC or IGF1R, but better in patients with high EZH2 or E2F1 patients than in patients with low EZH2 or E2F1.
Discussion
This study retrospectively evaluated the influence of the expression levels of 63 preselected genes (including three reference genes) on the outcomes of patients enrolled in the ACTS-GC. We found an association between high IGF1R or low AREG expression and poor prognosis. We concluded that IGF1R and AREG are prognostic, not predictive, markers of stage II/III gastric cancer.
IGF1R is a multifunctional tyrosine kinase receptor that is activated by its ligands, IGF1 and IGF2. IGF1R participates in several biological processes, including cell proliferation, differentiation, DNA repair, and prevention of apoptosis [14–17]. Aberrant activation of the IGF1/IGF1R axis has been associated with worse prognosis in many tumors, including breast, colorectal, laryngeal, myeloma, and prostate [18–20]. Data regarding IGF1R prognostic value in non-small cell lung cancer (NSCLC) are inconsistent [21–23]. Although relatively few gastric cancer cases have been evaluated, one report demonstrated that IGFIR overexpression in a primary tumor was correlated with increased lymph node metastasis, and that patients with low expression of both IGF1R and EGFR had significantly improved OS [24, 25]. In this study, IGF1R mRNA expression level was not correlated with tumor size, lymph node status, and staging of the tumors. IGF2, one of the ligands of IGF1R, could not be evaluated because its expression level was below the detection limit in less than 60 % of the samples. Previous papers that accounted for IGF1R analyzed relatively small numbers of samples in a retrospective manner, whereas the present study with its retrospective-prospective design enrolled 829 patients, showed the poor outcome of patients with high IGF1R expression, and successfully confirmed the prognostic value of this gene for gastric cancer. Furthermore, data from the publicly available database (GEO microarray data set) also supported the prognostic capability of IGF1R expression. Therefore, our results could encourage conducting further prospective studies to evaluate the IGF/IGFR axis.
AREG is a ligand for the epidermal growth factor receptor (EGFR), a transmembrane tyrosine kinase receptor that has a central role in regulating cell division and death [26]. AREG induces proliferative activities in various types of cells [27]. Recently, the effect of AREG on the prognosis and treatment efficacy of colorectal cancer patients receiving the anti-EGFR agent was investigated. High AREG or EREG expression identified a subgroup of KRAS wild-type patients who had a high probability of responding to EGFR inhibition [28]. The CO-17 study, which compared treatment with cetuximab and best supportive care (BSC) to BSC alone in patients with metastatic EGFR-positive colorectal cancer, revealed that EREG expression levels were positively correlated with cetuximab treatment efficacy [29]. Thus, AREG or EREG had a predictive value in patients treated with cetuximab. Interestingly, in patients with metastatic colorectal cancer receiving first-line chemotherapy without the anti-EGFR agent, high AREG or EREG expression significantly correlated with longer progression-free survival, and the positive prognostic value of high EREG was confirmed to be independent in a multivariate analysis [30]. Data regarding AREG prognostic values for NSCLC patients are inconsistent. Patients on the placebo arm with high AREG had statistically poorer OS than patients with low AREG, which remained significant in multivariate analysis, in the NCIC Clinical Trials Group BR.21 [31]. These discrepancies might depend on the difference of cancer type. We previously reported that patients with EGFR-positive tumors had worse survival than those with EGFR-negative tumors in the ACTS-GC biomarker study, when EGFR expression was evaluated by the immunohistochemical staining [4]. EGFR status had no relationship to AREG gene expression (data not shown). In addition, the prognostic values of AREG expression maintained in both patients with EGFR-positive (n = 75) and EGFR-negative (n = 752) tumors (data not shown). There have been few reports on AREG prognostic value in gastric cancer patients after surgery; the present ancestry study of the ACTS-GC is an important resource for evaluating the prognostic value of this gene.
The current study was limited by the following reasons. This study is for stage II and stage III patients, and this selection bias should be noticed to generalize our knowledge. The number of genes screened was relatively small. Additional useful candidate genes should be evaluated using archived cDNA from the present study in future investigations. Moreover, the correlation of gene expression according to mRNA measurement and protein levels should be further investigated using clinically feasible procedures such as immunohistochemical staining.
In conclusion, the current study provided compelling evidence that high IGF1R and low AREG expression were associated with poor prognosis after curative resection of stage II/III gastric cancer. There was no apparent interaction between S-1 and IGF1R or AREG status with respect to survival. These findings should contribute to the development of urgently required new targeted therapies for gastric cancer patients who are at high risk of relapse.
References
Shirasaka T, Shimamato Y, Ohshimo H, Yamaguchi M, Kato T, Yonekura K, et al. Development of a novel form of an oral 5-fluorouracil derivative (S-1) directed to the potentiation of the tumor selective cytotoxicity of 5-fluorouracil by two biochemical modulators. Anticancer Drugs. 1996;7:548–57.
Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, et al. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007;357:1810–20.
Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T, et al. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II/III gastric cancer: ACTS-GC. J Clin Oncol. 2011;29:4387–93.
Terashima M, Kitada K, Ochiai A, Ichikawa W, Kurahashi I, Sakuramoto S, et al. Impact of expression of human epidermal growth factor receptors EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin Cancer Res. 2012;18:5992–6000.
Sasako M, Terashima M, Ichikawa W, Ochiai A, Kitada K, Kurahashi I, et al. Impact of the expression of thymidylate synthase and dihydropyrimidine dehydrogenase genes on survival in stage II/III gastric cancer. Gastric Cancer. 2015;18:538–48.
Fareed KR, Kaye P, Soomro IN, Ilyas M, Martin S, Parsons SL, Madhusudan S. Biomarkers of response to therapy in oesophago-gastric cancer. Gut. 2009;58:127–143.
Ichikawa W. Prediction of clinical outcome of fluoropyrimidine-based chemotherapy for gastric cancer patients, in terms of the 5-fluorouracil metabolic pathway. Gastric Cancer. 2006;9(3):145–55.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;97:1180–4.
Ceppi P, Volante M, Novello S, Rapa I, Danenberg KD, Danenberg PV. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–25.
Bonanomi A, Kojic D, Giger B, Rickenbach Z, Jean-Richard-Dit-Bressel L, Berger C, et al. Quantitative cytokine gene expression in human tonsils at excision and during histoculture assessed by standardized and calibrated real-time PCR and novel data processing. J Immunol Methods. 2003;283:27–43.
Pérez S, Royo LJ, Astudillo A, Escudero D, Alvarez F, Rodríguez A, et al. Identifying the most suitable endogenous control for determining gene expression in hearts from organ donors. BMC Mol Biol. 2007;8:114.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034.
Lee J, Sohn I, Do IG, Kim KM, Park SH, Park JO, et al. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One. 2014;9(3):e90133.
Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505–18.
Li R, Pourpak A, Morris SW. Inhibition of the insulin-like growth factor-1 receptor (IGF1R) tyrosine kinase as a novel cancer therapy approach. J Med Chem. 2009;52:4981–5004.
Ludovini V, Bellezza G, Pistola L, Bianconi F, Di Carlo L, Sidoni A, et al. High coexpression of both insulin-like growth factor receptor-1 (IGFR-1) and epidermal growth factor receptor (EGFR) is associated with shorter disease-free survival in resected non-small-cell lung cancer patients. Ann Oncol. 2009;20:842–9.
Chitnis MM, Yuen JS, Protheroe AS, Pollak M, Macaulay VM. The type 1 insulin-like growth factor receptor pathway. Clin Cancer Res. 2008;14:6364–70.
Liao Y, Abel U, Grobholz R, Hermani A, Trojan L, Angel P, et al. Up-regulation of insulin-like growth factor axis components in human primary prostate cancer correlates with tumor grade. Hum Pathol. 2005;36:1186–96.
Mountzios G, Kostopoulos I, Kotoula V, Sfakianaki I, Fountzilas E, Markou K, et al. Insulin-like growth factor 1 receptor (IGF1R) expression and survival in operable squamous-cell laryngeal cancer. PLoS One. 2013;8:e54048.
Djavan B, Waldert M, Seitz C, Marberger M. Insulin-like growth factors and prostate cancer. World J Urol. 2001;19:225–33.
Cappuzzo F, Tallini G, Finocchiaro G, Wilson RS, Ligorio C, et al. Insulin-like growth factor receptor 1 (IGF1R) expression and survival in surgically resected non-small-cell lung cancer (NSCLC) patients. Ann Oncol. 2010;21:562–7.
Dziadziuszko R, Merrick DT, Witta SE, Mendoza AD, Szostakiewicz B, et al. Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non-small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol 2010;28:2174–2180.
Cappuzzo F, Toschi L, Tallini G, Ceresoli GL, Domenichini I, Bartolini S, et al. Insulin-like growth factor receptor 1 (IGFR-1) is significantly associated with longer survival in non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2006;17:1120–7.
Jiang Y, Wang L, Gong W, Wei D, Le X, Yao J, et al. A high expression level of insulin-like growth factor I receptor is associated with increased expression of transcription factor Sp1 and regional lymph node metastasis of human gastric cancer. Clin Exp Metastasis. 2004;21:755–64.
Matsubara J, Yamada Y, Nakajima TE, Kato K, Hamaguchi T, Shirao K, et al. Clinical significance of insulin-like growth factor type 1 receptor and epidermal growth factor receptor in patients with advanced gastric cancer. Oncology. 2008;74:76–83.
Di Fiore PP, Segatto O, Taylor WG, Aaronson SA, Pierce JH. EGF receptor and erbB-2 tyrosine kinase domains confer cell specificity for mitogenic signaling. Science. 1990;248(4951):79–83.
Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31(6):637–43.
Tabernero J, Cervantes A, Rivera F, Martinelli E, Rojo F, von Heydebreck A, et al. Pharmacogenomic and pharmacoproteomic studies of cetuximab in metastatic colorectal cancer: biomarker analysis of a phase I dose-escalation study. J Clin Oncol. 2010;28(7):1181–9.
Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–7.
Stahler A, Heinemann V, Giessen-Jung C, Crispin A, Schalhorn A, Stintzing S, et al. Influence of mRNA expression of epiregulin (EREG) and amphiregulin (AREG) on outcome of patients with metastatic colorectal cancer treated with 5-FU/LV plus irinotecan or irinotecan plus oxaliplatin as first-line treatment (FIRE 1-trial). Int J Cancer 2015;138:739–46.
Addison CL, Ding K, Zhao H, Le Maître A, Goss GD, Seymour L, et al. Plasma transforming growth factor alpha and amphiregulin protein levels in NCIC Clinical Trials Group BR. 21. J Clin Oncol 2010;28:5247–56.
Acknowledgments
We dedicate this manuscript to the memory of Prof. Tetsuro Kubota, who made a great contribution to the conception and design of this study. We thank the patients, their families, and the investigators from the 65 institutions for their cooperation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
I.K. and M.S. have acted in an advisory role for Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan). W.I., A.O., S.S., H.K., H.I., and M.S. have received research grants from Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan). W.I., M.T., A.O., S.S., H.K., T.S., H.I., and M.S. have received a speaker honorarium from Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan).
Ethical standards
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. The protocol used for the current biomarker study was approved by the ethics committee of the Japanese Gastric Cancer Association and the institutional review board of each participating hospital.
Funding
This study was funded by Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan). We did not have any grant to support this study.
Additional information
For the ACTS-GC Group.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ichikawa, W., Terashima, M., Ochiai, A. et al. Impact of insulin-like growth factor-1 receptor and amphiregulin expression on survival in patients with stage II/III gastric cancer enrolled in the Adjuvant Chemotherapy Trial of S-1 for Gastric Cancer. Gastric Cancer 20, 263–273 (2017). https://doi.org/10.1007/s10120-016-0600-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-016-0600-x