Abstract
In data science and content-based retrieval, we find many domain-specific techniques that employ a data processing pipeline with two fundamental steps. First, data entities are represented by some visualizations, while in the second step, the visualizations are used with a machine learning model to extract deep features. Deep convolutional neural networks (DCNN) became the standard and reliable choice. The purpose of using DCNN is either a specific classification task or just a deep feature representation of visual data for additional processing (e.g., similarity search). Whereas the deep feature extraction is a domain-agnostic step in the pipeline (inference of an arbitrary visual input), the visualization design itself is domain-dependent and ad hoc for every use case. In this paper, we survey and analyze many instances of data visualizations used with deep learning models (mostly DCNN) for domain-specific tasks. Based on the analysis, we synthesize a taxonomy that provides a systematic overview of visualization techniques suitable for usage with the models. The aim of the taxonomy is to enable the future generalization of the visualization design process to become completely domain-agnostic, leading to the automation of the entire feature extraction pipeline. As the ultimate goal, such an automated pipeline could lead to universal deep feature data representations for content-based retrieval.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
With the advent of the big data era, we observed a massive explosion of volume and complexity of data. The traditional data engineering prism divides the data into structured (e.g., relational) and unstructured (text documents, multimedia, time series, sensory data [85]). However, when it comes to retrieving data, this structured/unstructured dichotomy may not be very relevant. Instead, the critical property of the data is its understandability (or interpretability) by a human user. Understandable data require description at a high level of semantics that could be used by a human user (e.g., data scientist, system designer) for retrieval and other tasks of data management.
The mentioned increasing volume and complexity of data are often connected with the decreasing semantic level of data description. This lack of high-level semantics is a problem not just for unstructured data but also for low-level structured data. For instance, in the medical domain, multi-modal patient data (as shown in Fig. 1) can include images (such as MRI scans of the brain), multidimensional time series (like EEG recordings), free text (patient anamnesis), and various numerical measurements stored in a relational database (biomarkers). Without human-curated metadata, all these raw data suffer from low-level semantics of the data representation. Even accessing relational data, represented by the table with biomarkers, can be challenging through conventional methods like SQL. The table contains dozens of numeric attributes forming multidimensional feature vectors rather than semantic attributes that can be used within the SQL SELECT clause. Therefore, the table’s low-level semantic structure is comparable to unstructured data like time series, consisting of a sequence of values or multimedia, where an image is a matrix of color-pixel values.
Defining a versatile and structured retrieval model for complex low-level data, whether structured or unstructured, is challenging (analogous to SQL for high-level relational data). Instead, these data are mostly retrieved using content-based similarity search models (metric [31, 99], or non-metric [75, 77]), where data instances are represented by feature descriptors, typically high-dimensional vectors, that are comparable by a similarity function. This mechanism allows for designing a universal retrieval engine, where similarity queries are executed in the query-by-example manner. The query by example is an elegant way to protect users from needing to use low-level query descriptions. For example, in content-based image retrieval, the user provides an example image of a dog and searches for other dog images that are most similar to the example. Hence, the user is not required to understand the internal data representation (as they would in the case of semantic relational data).
The retrieval model based on similarity search provides a unified solution for complex low-semantic data retrieval. However, it does not include the extraction of data features into the descriptors. This domain-specific part of the model must be addressed individually for each use case. Obviously, a possible extension of the similarity-based retrieval model to also automate descriptor design and feature extraction would be a major breakthrough in many fields of data engineering. To achieve such an ultimate goal, we could try to combine techniques from two successful research areas—machine learning and data visualization.
In the field of machine learning, many models of visual-semantic descriptors for visual data (video, images) based on deep convolutional neural networks (DCNN) have been developed. Usually, these models greatly outperform models based on hand-engineered features [27]. Additionally, DCNN models have demonstrated a generalization effect, producing semantic embeddings (deep-feature vectors) that work well outside their original training domain [19, 56]. For instance, a DCNN model could represent images of veteran cars even when trained on an image collection with no veteran car images in it. This generalization effect might set DCNN models in the role of universal deep-feature extractors for complex data. However, a large share of data is non-visual in its primary representation, so that “visual” DCNN models cannot be directly used with them (e.g., the mentioned medical data). However, this limitation could be addressed by the introduction of a systematic visualization model for complex low-level data. Data represented by suitable visualizations (i.e., by artificial images) could then be used with DCNNs, similarly to the “veteran car” example.
The use of visualization approaches goes beyond facilitating the processing of data by visual machine learning models like DCNNs. It also has significant value in fields such as human-computer interaction (HCI) and information visualization, as it may help users to understand data not only analytically, but also cognitively / intuitively. This intuitive aspect is particularly important for complex data that is difficult to interpret through purely analytical means. Last but not least, visualization of data at different abstraction levels helps a human user / specialist understand the semantics of the entire data processing pipeline and thus addresses the hot problem of today: the explainability/interpretability of AI [67, 98].
1.1 Paper contributions
In our previous work [76], we have sketched a transfer-learning framework where a pretrained DCNN model is generalized to a universal feature extractor (or classifier) for any kind of data that can be suitably visualized. In this survey, we elaborate more on the most challenging part of the framework—the visualization of any kind of data. To be successful, the visualization design should respect some premises that allow mapping non-visual features of domain-specific data into visual elements already recognized in the models (we further discuss the premises in Sect. 3.1). Specifically, the main contributions of this paper are as follows.
-
We review and analyze numerous domain-specific cases where DCNN (or related) models were used in the role of feature extractors or classifiers, transforming visualizations of data instances into deep-feature representations.
-
Based on the analysis of the reviewed cases, we propose a data visualization taxonomy for heterogeneous data and formulate premises for DCNN-compliant visualizations. The taxonomy gives a systematic overview and possibly also guidelines for visual representations of heterogeneous data that are suitable for DCNN models. We also map the particular visualization models used in the reviewed cases into the proposed taxonomy.
-
In general, the review reveals that non-visual data can often be visualized by artificial images and successfully used in machine learning models.
2 Background
In this section, we summarize the basics of deep convolutional neural networks and visualization taxonomies, representing the two crucial areas used in the above-mentioned framework.
2.1 Deep convolutional neural networks
A decade ago, deep convolutional neural networks (DCNN) became the state-of-the-art solution for many problems in computer vision, including classification and retrieval. Moreover, from the wider perspective of data engineering, DCNN models could be utilized beyond a closely defined task, such as classification. They can extract deep features from input images, formed by the activations of neurons at a specific network layer, which are suitable for similarity search and other forms of content-based retrieval, analytics, and management [91]. For example, the pioneering AlexNet model [39] contained five convolutional layers, three fully connected layers, one class probability layer, and several additional types of layers. One image can thus be represented by deep-feature vectors at different levels of abstraction (from local, visual to global, semantic). The development of DCNN architectures has continued at a swift pace through the last years, producing GoogleNet [83], VGGNet [74], ResNet [30], SENet [32], EfficientNet [84], and others.
It has been shown [24] that deep features obtained from a generic pretrained DCNN model such as ResNet work well when applied to domains, datasets, and recognition tasks different from those they were trained on. This fact is crucial as we aim at re-using generic DCNN models in various domains to avoid costly model training for every domain.
2.1.1 Fine-tuning a DCNN model
Although DCNN models are being designed and trained for a particular task, such as domain-specific classification, they could also be re-purposed for other domains. The process of reusing a model in a different domain is known as transfer learning [58]. The most common and straightforward method of transfer learning is fine-tuning a pretrained model. This involves replacing the neurons in the model’s classification layer with a new set representing the target domain’s classes. The model is then trained further with a labeled dataset from the target domain. The advantage over developing a new model design is that the target-domain training dataset could be much smaller than the dataset used to train the original model or the dataset needed to train a new model from scratch.
2.1.2 Non-CNN models
Even though DCNNs are considered state-of-the-art in computer vision, similar to natural language processing, models based purely on attention have been developed recently. Specifically, Vision Transformer (ViT) [20] is a transformer model in which the image is divided into patches and treated as an image sequence in the same way as language transformers treat a sequence of tokens. Usage of such a model violates some of the premises presented in this study (see Sect. 3.1) because transformer models do not use any convolutional blocks. On the other hand, similarly to DCNN, also ViT can be used in transfer learning scenarios. With an increasing amount of pretrained ViT models, transformers might become a viable alternative to the deep-feature representation approaches surveyed here. However, as ViTs are a very recent development, none of the approaches surveyed in this article were based on this architecture. Therefore, only convolution-based models are considered further on.
2.2 Visualization taxonomies
Let us now briefly overview the existing generic data visualization taxonomies that served as the basis for our taxonomy capturing data and representations suitable to use with DCNN models (see Sect. 3).
Information visualization taxonomical frameworks revolve around different aspects of the visualization process, such as visualization technique, visualization task or interactivity. As visualization is the process of mapping data to a visual representation, every taxonomy includes the data aspect, which is then associated with one or more visualization aspects. Most work on visualization taxonomies can be traced back to the work of Bertin [42], who describes how different types of data map to different types of graphical marks and how these can be aggregated into complex visualizations, i.e., visualization techniques. Bertin decomposes the data into values and structure, where values are associated with simple data types, while the structure is associated with relations. Bertin’s taxonomy was later extended or modified by others such as Tweedie [87], who added transformation of the raw data into Bertin’s taxonomy, Mackinlay [48], who put forward the idea that graphical presentations are sentences of graphical languages and codified the visualization design process, or Tory and Moller [86], who relaxed the dependence on data type by categorizing the visualization techniques based on models of data rather than the underlying data themselves.
Visual techniques can be classified according to different criteria. Lohse et al. [45] created a hierarchical classification of 60 visual techniques (representations in the language of the paper) based on the responses of a cohort of users. Later, Gleicher et al. [28] argued that visualization techniques should be studied in general, independently of what objects they contain, and classified the design into three categories that can be combined: juxtaposition (separation), superposition (overlay), and an explicit representation of relationships. A different view was offered by Rodrigues et al. [65], who classified techniques based on perceptual-cognitive perspective. Along these rather general classification approaches, more specific taxonomies have been developed to classify specific visualization techniques such as treemaps [68].
Another body of work has been done on the visualization task taxonomies starting with Shneiderman’s task by data type taxonomy with seven data types (one-, two-, three-dimensional data, temporal and multi-dimensional data, and tree and network data) and seven tasks (overview, zoom, filter, details-on-demand, relate, history, and extracts) [72]. Visual task taxonomies have been later extended to other domains such as temporal [43], graph [2, 43], or multivariate [57] data analysis. For more on visualization task taxonomies, we refer to an extensive review of the literature [69].
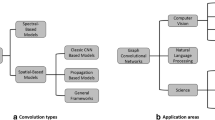
All of the taxonomies mentioned above describe a single aspect of the information visualization process. The individual aspects can be combined into multi-aspect taxonomies, a notable example of which is the one created by Keim [37], who puts together the data, visualization technique (display mode in the language of the taxonomy), and task (interaction and distortion in the language of the taxonomy), forming an orthogonal taxonomy. The data types include 1D, 2D, multi-dimensional, text/web, hierarchies/graphs, and algorithm/software. Visualization techniques are taken from a taxonomy previously introduced by Keim [38] and include standard 2D/3D display, geometrically transformed display, iconic display, dense pixel display, and stacked display. Finally, the interactivity aspect includes projection, filtering, zooming, distortion, linking, and brushing. The individual aspects are combined into an orthogonal taxonomical system (see Fig. 2a). Although thinking of the individual aspects as orthogonal feels natural, it can be a bit misleading since not all combinations of values are valid, as noticed by Chengzhi et al. [11]. This snag is not present in the work of Pfitzner et al. [59], who builds on Chi [12] by introducing a taxonomical framework where the aspects of the visualization process are mutually referenced (see Fig. 2b) but do not form a complete Cartesian space.
3 Visualization taxonomy for DCNN-based data representations
The analyzed approaches (see Sect. 4) proposed a plethora of different visualization techniques to transform originally non-visual data into representations suitable for DCNN-based post-processing. However, the abundance of techniques necessitates some organization over them. To address this, we propose a new taxonomy oriented on the data types and the structuredness of data visualizations. Although our work is inspired by existing related works on visual taxonomies (see the previous section), we argue that none of the existing taxonomies adequately capture the specifics of data and visualization techniques used in the DCNN approaches.
The existing taxonomies either aim to characterize the entire information visualization space by capturing many aspects of the visualization process, or to characterize the full breadth of an individual aspect (e.g., all data types). Neither of those approaches is suitable in our use-case as some aspects, such as the visual task, are not applicable at all, while other aspects, such as the visualization techniques, are applicable only partially (for instance, 3D displays do not make sense for 2D DCNNs). Therefore, we propose a two-aspect taxonomy, in which data types are mapped to visualization techniques (see Fig. 3). Our set of visualization techniques resembles those defined by Keim [38]. On top of the visualization techniques, we also introduce a hierarchy similar to the data types hierarchy using a binary classification tree diagram proposed by Lohse et al. [45]. However, unlike Keim, we do not treat the taxonomy as a Cartesian space, but rather as a bipartite graph, where relevant parts of each aspect are connected.
The first aspect of our taxonomy aims at describing visualization techniques in terms of their structuredness, i.e., the granularity and the nature of visualized entities. On the top level, we distinguish unstructured, structured, and semi-structured visualizations. In unstructured visualizations, the shape of displayed entities bears no further meaning. Essentially, colored pixels or blobs of a single color are displayed. This corresponds to the dense pixel display as described in [37]. Structured visualizations, on the other hand, aim to plot more complex entities such as symbols or graph elements. The shape of depicted entities has an intended meaning, which is usually human-understandable. We further distinguish semi-structured representations in the intersection of both previous variants. Variants of semi-structured representations may correspond, e.g., to colored overlays over structured objects or visual transformations of structured objects. On the next level, we aim to further distinguish the types of displayed objects, i.e., 2D graph elements or symbols of some alphabet for structured representations, and the organization of color blobs, i.e., arbitrary, fixed or trained ordering of elements for unstructured representations.
The suitability of a visualization technique depends heavily on the nature of the source data and any potential transformations that may be applied to them. The most significant phenomenon that affects the applicability of a visualization technique appears to be the knowledge of a mutual relationship between individual features describing an object. Therefore, in the second aspect of the proposed taxonomy, we differentiate between tabular, sequential, and graph source data.
By tabular data, we understand a set of n objects \(X=\{x_1, x_2, \dots , x_n\}\), where each \(x_i, i\in \{1,2,\dots , n\}\) is a vector of m features. Such data can be represented as a table with objects as rows and features as columns. Unlike sequential or graph data, no ordering or hierarchy of features is defined on the featuresFootnote 1
By sequential data (e.g., time series or a stream of text), we understand a set of n objects \(X=\{x_1, x_2, \dots , x_n\}\), where each \(x_i, i\in \{1,2,\dots , n\}\) is a collection of linearly ordered observations \(x_{i_t}\), \(t\in \{1, 2, \dots , m\}\). The sequence index t can be viewed, e.g., as time, in which case X represents a time series. Sequential data can be further classified as univariate (single-dimensional) or multivariate (multi-dimensional) depending on whether \(x_{i_t}\) is a scalar or a vector of features.Footnote 2
Finally, by graph data we understand a set of n objects \(X=\{x_1,x_2,\dots ,x_n\}\), where each \(x_i, i\in \{1,2,\dots , n\}\) is a graph, that is, an ordered pair \(G_i=(V_i,E_i)\), where \(V_i\) represents a set of (possibly labeled) vertices \(V=\{v_{i_1}, v_{i_2}, \dots , v_{i_n}\}\) and \(E_i\) is the set of edges (possibly oriented and with associated edge types) such that \(E_i \subseteq \{\{v_{i_k},v_{i_l}\} \mid v_{i_k},v_{i_l} \in V_i, v_{i_k} \ne v_{i_l} \} \). As such, more complex structures or hierarchies can be disclosed.
Schema of the proposed taxonomy. Top: considered types of source data, bottom: structuredness of data visualization (with example visualizations). Edges represent approaches used in the reviewed literature combining a particular data type and the structuredness of a visualization technique. Note that for each edge, we describe transformations of the source data needed for a particular visualization type. If multiple such transformations were present in the reviewed papers, we depict the simplest one. Rare visualization variants (i.e., only one reference found) are denoted with a dashed line, while more common ones with a solid line. Displayed visualization examples are reprinted from [4, 5, 9, 23, 81, 90]
In Fig. 3, we depict the relationship between the source data types and the applied visualization techniques. Note that we only depict links based on the reviewed papers—some additional connections are theoretically plausible.Footnote 3
3.1 Premises on DCNN-compliant visualizations
The data visualizations studied in this work should be interpretable by both a human and a machine (a DCNN model in our case). At this point, we would like to discuss some premises leading to visualizations suitable for machine interpretability. The premises are tightly bound to the general architecture of DCNN models. DCNN architectures were inspired by the human cognition system and usually consist of several vision layers (convolutional, pooling) followed by several fully-connected layers. Technically, a trained DCNN model is a system of learned feature detectors at different semantic levels. Training data for the mainstream DCNN models are typically labeled photographs of general real-world scenes (such as the ImageNet image database [17]).
Besides the primary DCNN task (such as image classification), an inference of a visual input also provides the deep-feature vector (image embedding). In this work, we study DCNN models as feature extractors for visualized data, that is, for artificial images (not real-world photography anymore). Hence, as DCNN models were originally developed for real-world photography, their repurposing for artificial visualizations should respect some implicit premises to keep the visual properties of visualizations close to the original (photographic) training data. We define the premises for DCNN-compliant visualizations as follows:
-
1.
Local compliance. Real-world photographs exhibit a great degree of smoothness in color and brightness intensities within small regions of pixels. As image convolution is the key operation in DCNNs, the visual properties should be robust to quantifying the effects of convolution. As a consequence, key properties of the data entities should be visualized by larger contiguous pixel patches, not by single discrete pixels with precise positioning. Alternatively, closely related features should also maintain spatial proximity in the visualization. Also, the pixels/patches describing local properties should be invariant to (some) affine transformations, meaning that their absolute positions, scale, and rotation should not significantly affect the visualization.
-
2.
Global compliance. At deeper layers of DCNN models (either convolutional or fully-connected), the visual properties common in real photography are more global, more delicate, and more abstract. They could represent a pattern in the image layout (e.g., vertical arrangement of artifacts) or even very abstract quality (e.g., romantic style photography [35]).
-
3.
Descriptiveness. As with any other data descriptor, the visualization should be descriptive enough to distinguish between similar and non-similar data entities. In the case of a visual descriptor (artificial image), relevant/related data entities should be visually similar and vice versa. This assumption is consistent with the goal of human- and machine-interpretable data visualizations.
4 Mapping of domain cases to the taxonomy
In this section, we provide a review of approaches that utilize data visualization as a means to employ DCNNs to solve some domain-specific tasks. These approaches are mapped into the previously introduced taxonomy, but we also aim at highlighting additional interesting details of the respective approaches. The review is organized into subsections corresponding to the visualization types as described in Fig. 3.
Surveyed approaches are also listed in Tables 1, 2, 3, 4 and 5. Here, approaches are grouped with respect to the structuredness of data visualization, but additional information is also available. Namely, we depict the type of input data, utilized data transformations, whether some form of transfer learning was utilized in the underlying neural network, the target domain of the work, and the publication year. Note that if multiple approaches for data transformation were proposed in respective papers, we describe the simplest variant. Regarding transfer learning, we consider three situations as illustrated in Fig. 4. If the underlying network was trained from scratch without any transfer learning, we refer to it as “Fully trained”. If the network was originally trained on another domain (ImageNet classification in most cases), but then a substantial portion of the network was fine-tuned for the current task, we denote it as “Fine-tuned”. Finally, the network is denoted as “Pretrained” if a substantial portion of it was frozen, and we can consider the output of the frozen part as an embedding of the input visualization. Finally, for the sake of context, we also reviewed several (mostly older) approaches using hand-crafted visual features such as SIFT or MPEG-7 instead of DCNN. These are referred to as “not DCNN” in the tables.
Note that the distinction between “fine-tuned” and “pretrained” approaches is somewhat fuzzy. For instance, it was quite common among reviewed approaches that the whole convolution and pooling section of the pretrained network was kept intact, and modifications were only made to the fully connected layers (in most cases, these layers were simply replaced by new ones). One can consider this as a fine-tuning because the network was adapted to the new task. However, we rather denote this as a pretrained case because the frozen part of the network can be considered as an independent network delivering a meaningful embedding of the visualization. This embedding can then be supplied as source data to any shallow machine learning algorithm trained on the actual task. By this, we unite various machine learning approaches (including shallow neural networks appended to the embeddings calculation) and distinguish this case from making more substantial changes (i.e., within the convolution and pooling section) of the DCNN model itself.
Visual legend to the “Training” column of Tables 1, 2, 3, 4 and 5. Typically, DCNN networks consist of the input layer followed by numerous convolution and pooling layers. Then, its output is combined via several fully connected (FC) layers, and finally, an output prediction is produced. Note that we only denote approaches as “fine-tuned” if the training on the current domain also modifies weights in the convolution & pooling section of the network. We argue that otherwise (i.e., when only FC layers are modified), this is equivalent to using a pretrained network followed by some shallow machine learning (ML) algorithm
4.1 Structured visualization
First, we review structured visualization methods (see Table 1), where the visual representation of the underlying data is easiest to understand by a human observer, i.e., graphs and symbols.
4.1.1 Graphs
Chemoinformatics is a typical domain where graphs are natural representations of the underlying entities. Graphs usually represent molecular structures with atoms corresponding to nodes and covalently bonded atoms as edges connecting the corresponding nodes. As the 2D drawings of these so-called molecular graphs are standardized, there exist DCNN applications over such drawings. Probably the first approach applying DCNN to molecular graph drawings was Chemception [29], motivated by Google’s Inception-ResNet, where molecular graphs were treated simply as images with no explicit chemical knowledge provided to the network (Fig. 5a). Chemception was successfully applied to both binary classification (activity and toxicity of a compound) and regression (free energy of solvation). A similar approach was used in the DeepScreen method [64], where the authors built a large-scale drug-target interaction system using only 2D images of known drugs and successfully used it to identify new targets for existing drugs. Another work in which DCNNs over molecular graph drawings were used for toxicity prediction was Toxic Colors [23]. In this case, the molecular graph drawings were further augmented by coloring certain areas based on the electrical charge at that position (Fig. 5b).
4.1.2 Layout of symbols
The pioneering work in the field of direct visualization of textual data as images (i.e., the layout of symbols from the text’s alphabet) is the so-called SuperCharacter method [80] (see Fig. 6a). In the method, an image containing the symbols of the given text (its initial part of a fixed length, respectively) was created and plotted one by one in a natural order. The resulting image was then passed to a DCNN focusing on sentiment analysis. The method was motivated by a holistic view of human perception, that is, the ability of humans to “screen through” text to gain some knowledge about it. Another indirect evidence supporting the viability of this approach is the fact that various deep architectures (e.g., CLIP [61], which maps free text and images to a joint latent space) are able to perform well in rather specific tasks such as optical character recognition (OCR). This suggests that they are capable of distinguishing between arbitrary language symbols, although they are not specifically designed or trained to do so. On the other hand, using a visual representation of textual data introduces an additional layer of complexity and possibly an additional source of errors. While the work was rather thoroughly evaluated over approaches available at that time, a comparison with more state-of-the-art language models (e.g., BERT [18] or GPT3 [8]) is missing.
The original SuperCharacter method utilized the Chinese language. Some modifications (compact layout of individual words, importance-based scaling) were proposed to handle English texts [81] (see Fig. 6b) and tabular data [82] (see Fig. 6c). A similar approach was also proposed to classify longer texts, e.g., financial reports [13].
4.2 Semi-structured visualization
The main feature of semi-structured visualizations (see Table 2) is that they combine both structured and unstructured components. Although semi-structured visualizations were rare in the reviewed papers, we consider them to be of great potential, as the resulting visualizations tend to fulfill the DCNN-compliance premises to a great degree.
The main idea behind [9] was to encode the data as a transformation of some source image. The authors focused on the problem of gene expression data classification and utilized the following pipeline. First, some source (real-world) image is selected and then the convolution operation is applied on it. The convolution kernel values are derived from the feature values describing each object, therefore, resulting in unique modifications of the source image per object. Figure 7a illustrates this approach.
One possible generalization of the aforementioned approach is the usage of generative adversarial networks (GANs) such as in [79]. Instead of transforming a single image, GANs generate artificial images while maintaining the distribution of the train database of (structured) images. Image generation is controlled by the seed vector which, to some extent, propagates the embedding’s similarity (i.e., similar seed vectors correspond to similar output images). Sreehari [79] exploited this property as follows. They utilized a pretrained GAN model (trained on the MNIST handwritten digits), while the seed vectors corresponded to the (encoded) features of the respective objects. Figure 7b visualize their pipeline.
4.3 Unstructured visualization
The common characteristic of methods classified as unstructured visualizations is the assignment of feature values to a rectangular areaFootnote 4 (typically a pixel or a square). Thus, the final visualization effectively corresponds to a heatmap. The individual approaches then differ in the way in which the features are mutually positioned, i.e., in their mutual order. We distinguish situations where features are placed in arbitrary order, the order is decided based on some external knowledge (fixed order), or the order is learned based on the characteristics of the dataset (trained order).
4.3.1 Arbitrary order
The arbitrary ordering (see Table 3) represents the most simple case of unstructured visualization. The visualization pipeline, in the simplest form, maps each feature to a pixel, and the pixel’s brightness/color reflects the feature value. Due to arbitrary ordering of the features, there is no relation between neighboring areas of the output image, which to some extent violates the local compliance premise.Footnote 5
This approach was used to classify network data from the Internet of Medical Things [94]. The value of each feature was linearly normalized to the interval [0, 255]. Subsequently, the features were mapped to pixels in an image of size \(\sqrt{M}\), where M was the number of features. A similar approach was used to predict neonatal sepsis [4], or for diabetes classification [34], where the normalization was based on the sigmoid function.
Several methods tried to extend the simplest feature-to-pixel approach. In the credit scoring area (financial domain), the Weight of Evidence transformation technique was utilized to bin the values of both categorical and numerical features [15]. The output image was a visualization of a sparse binary matrix of size \(N \times M\), where N was the number of bins and M was the number of features. Another approach proposed the use of feature importance to ensure that more essential features covered more area in the output image [78]. A different method to map features to pixels was proposed by Xu et al. [95]. In order to capture the interactions between features, the output visualization was produced directly by the \(\vec {x}\vec {x}^T\) transformation, where \(\vec {x}\) was a vector composed of features linearly normalized to the interval [0, 1]. This method was used to predict the mechanical properties of hot-rolled alloy steel.
4.3.2 Fixed order
As compared to the previous category, fixed order approaches (see Table 4) utilize some externally defined ordering or hierarchy of the displayed features. The most typical example is the visualization of time series, where one of the visualization axes represents a temporal sequence of values for some particular feature. Nonetheless, more complex visualization pipelines involve, for example, discrete signal transformation [3, 93] or complex feature hierarchy [46]. As the type of this external knowledge heavily impacted the proposed visualizations, this section is further divided based on the utilized data types (i.e., univariate, multivariate, and hierarchical).
Univariate sequential data
In this case, each object is described by a single modality, and the values are linearly ordered. Some examples of univariate sequential data include electrocardiograms (ECGs) [3, 53, 55, 66], audio signals [90], or a stream of bytes representing a binary code [6, 54, 88].
Probably the most straightforward visualization is directly encoding each value as a color (e.g., grayscale) and then serializing them into an image of desired dimensions. This approach was utilized in [55] for the classification of ECG. Each time series was trimmed to exactly 1024 values and then serialized to 32 rows and 32 columns in a line-first fashion.
Another domain using a similar type of transformation is binary code visualization for malware detection and defect prediction. Nataraj et al. [54] presented the idea of visualizing binary code as a sequence of grayscale pixels, where each byte is represented by one pixel. The sequence was then wrapped w.r.t. some predefined line width. Recently, Bozkir et al. [6] compared visualizations w.r.t. different line width definitions on a malware classification task. Results showed the benefits of using wider (e.g., 4096px wide) rather than longer (in terms of the image height) representations. Given the applied re-sizing, this essentially indicates the possibility to compress the information in subsequent bytes. Subsequent research differs by targeting additional deep neural networks or their combinations [14, 41, 49, 97], improving image information by introducing color channels [51, 88], or changing the ordering of individual pixels in images [63, 89].
Byte mapping visualization was also used for some related tasks. One notable example is \(\alpha \)Diff [44] aiming to detect similarity in cross-version binary codes. Note that in order to adapt to the task (code similarity vs. malware classification), some modifications were proposed in the image processing pipeline, e.g., replacing the plain DCNN with a Siamese network [7]. The example of \(\alpha \)Diff image is shown in Fig. 8.
Example of binary files comparison using \(\alpha \)Diff (generated as described in [44])
Another straightforward way of visualizing software was proposed by Chen et al. [10] for software defect detection task. In this case, the source code, rather than the compiled code, was visualized. In particular, three subsequent ASCII characters were encoded into one color pixel w.r.t. R, G, and B channels. Additionally, an image data augmentation method was proposed based on the permutation of the order of a pixel’s components.
The so far discussed approaches aimed to encode feature values directly as color blobs. Nonetheless, several authors further experimented with various forms of feature quantization.
In the bioinformatics domain, a histogram-based visualization was applied to sequential data in the metagenome classification task [1]. The authors split a metagenomic sequence into subsequences with a fixed size and stride and created a histogram based on subsequence co-occurrences. In the output visualization, the intensity of the pixel at position \(p_{i,j}\) was determined by a relative frequency of subsequences \(x_i\) and \(x_j\) in the input sample. However, this histogram was not used directly, but was one of the inputs of the attention layer in the classification pipeline.
Examples of visualizations based on time-frequency analysis of univariate time-series (ECG signal), reprinted from [53]: short-time Fourier transformation (STFT), continuous wavelet transformation (CWT) and Wavelet synchrosqueezing transform (WSST)
In the time series domain, multiple authors applied some form of time-frequency analysis, such as Fourier transformation [40, 66, 90] or continuous wavelet transformation [3, 33] (see Fig. 9). A common denominator of the described approaches is that subsequent chunks of the univariate data are transformed into a vector of features representing the intensity of the signal w.r.t. increasing frequency. These vectors are mapped into colors and then stacked along the time axis to form a 2D image. Therefore, one axis of the visualization represents time, while the other represents the frequency characteristics of the signal. In some cases, e.g., in [53] for ECG classification, the output of DCNN is combined with additional pretrained feature extractors. Then, some shallow ML algorithm is trained on top of the concatenated features. This may be a viable option if some meaningful features are easy to define. However, this usually requires substantial domain knowledge and as such, the approach is difficult to transfer to another domain.
In addition to time-frequency analysis, another option, phase-space analysis, was proposed [16]. First, a 2D phase space was created by assigning a time delay embedding to each point in the time-series data. Then the entire time-series data was transformed into a trajectory in this 2D phase space. On the basis of this trajectory, the recurrence plot was constructed reflecting the distance of states in the phase space, as shown in Fig. 10.
Time-series recurrence plot construction (reprinted from [16])
Multivariate sequential data
The multivariate sequential data, in contrast to univariate data, produce an additional axis that needs to be somehow accommodated in the visualizations. A straightforward approach (utilized, for example, in [21], see Fig. 11a) is to create univariate representations for each dimension (e.g., for each electrode in EEG or for each skeleton joint in gait recognition). Then, these univariate visualizations are stacked along one axis, while the temporal dependence is visualized along the other axis of the resulting image.
A similar approach [60] was proposed to classify fair and cheating players in online gaming using their keystroke dynamics. Player actions were converted into several independent time series, and then, using several consecutive DCNNs, behavioral patterns between fair and cheating players were recognized. Quite important here was the fact that the visualizations were actually two (see Fig. 11b). One visualization was used for DCNN, which expects several 1D time series in which the color of each pixel represents the value at a given time. The second visualization was more suitable from a human understandability perspective, so the time series was represented as a series of standard 2D curves where on the y axis we find the value at a given time x. The transformation from the human perspective into the DCNN perspective is shown in Fig. 11b.
Several reviewed approaches utilized an intermediate transformation step. This was used to visualize individual dimensions of the multivariate time series, and only then these partial visualizations were glued together. Some examples of such transformations are the usage of Gramian Angular Summation/Difference Field (GAF) [96], Markov Transition Field (MTF) [96] or Short time Fourier transformation [93].
Aside from stacking visualizations of individual dimensions, several alternatives were proposed to handle the problem of visualizing multiple features simultaneously.
One of the simple approaches was to project only the most relevant dimensions into a shared space (e.g., as color dimensions). This approach was utilized in [93] for EEG classification. The authors selected the three most relevant electrodes, whose output was encoded as R, G, and B channels of the resulting image via short-time Fourier transformation. Note that similar mapping was also applied in [21] to the problem of gait recognition. However, in this case, color mapping was applied to the 3D spatial coordinates of individual skeletal joints.
In [26], authors focused on classifying several states of epileptic EEG. In order to do so, they separated the time series into short sequences of a fixed length (4 s) and focused on the energy variation along various frequencies. The power spectrum density analysis was applied to the whole sequence of the signal, so each 4sec sequence resulted in one image, where individual electrodes were stacked along one axis, while the power spectrum density for increasing frequencies was depicted along the other axis. Essentially, the temporal dimension of the time series was abstracted from the visualization and replaced by a sequence of images representing the same object at different times. A similar approach was also applied to the fMRI data [25], where each data point corresponds to a four-dimensional spatiotemporal coordinate. In this case, two of the spatial dimensions were preserved within each image, while the remaining two dimensions were utilized to create the sequence.
Hierarchical data
The fixed order of the visualized features may follow more complex data organization than linear ordering (as in univariate or multivariate sequential data). In [46], the authors utilized TreeMap [71], a 2D space-filling method for hierarchical visualization, to organize gene expression data of individual samples in such a way that molecular features related in function are spatially adjacent. For this purpose, a predefined taxonomy was established, capturing the functional hierarchy of biological objects—genes, in this case (see Fig. 12). To encode a sample by an image, the expression of each gene was encoded by the color intensity of a position corresponding to that particular gene. DCNN was then trained on these images to predict the grades of different types of cancer. Moreover, identified informative features of the network could be used to reveal contributory genes and provide the basis for further discoveries.
Treemap representation of functional hierarchy of genes (reprinted from [46])
4.3.3 Trained order
Trained order visualizations (see Table 5) aim to mitigate the main drawback of arbitrary order visualizations, that is, violation of the local compliance premise. In particular, trained order visualizations embed features into the 2D space, so that similar features are clustered together. The effect of such embedding is similar as in the case of the fixed order representations, i.e., constructing stronger clusters of the signal.Footnote 6
All surveyed trained order approaches learn how to assign features into fixed pixel positions (i.e., features are re-ordered, but mapping to a certain position within the image is then fixed). The methods in this class differ mainly in the way they approach the feature-to-pixel mapping process.
The most simple trained order methods are based on direct feature-to-pixel techniques presented in Sect. 4.3.1 with the addition of an ordering method that arranges feature representations in the output visualization. A straightforward ordering method employing feature similarity based on Pearson’s correlation was used by Feng et al. for network intrusion classification [22]. An ordering method based on Pearson’s correlation coefficient was also used in the visualization pipeline proposed by Sreehari et al. [79]. In the first step, the features were ordered with respect to their similarity. Subsequently, the number of pixels in the output image assigned to a particular feature was determined by its importance. Each pixel value was drawn from a normal distribution with a mean equal to the feature value and variance equal to the feature variance across all observations. The last step of the visualization pipeline was the application of a zero-mean Gaussian kernel on the final image to emphasize interactions of visualized features. Nevertheless, neither of the aforementioned methods specified the exact algorithm they use for feature ordering.
Feature similarity was also used in visualizations of multivariate time-series patient data for the mortality prediction [50]. The proposed method combined the stacking of individual feature visualizations along the horizontal axis, which represented the time dimension, with the spatial arrangement of the features in the vertical axis with respect to their correlations. In this case, the R package corrplot was used to determine the exact feature ordering.
In the bioinformatics domain, the first trained order method we are aware of was DeepInsight [70]. In DeepInsight, the authors treat features as objects in n-dimensional space and used dimension reduction techniques such as tSNE [47] to embed the features in the 2D space. Once the location of features is determined, the convex hull algorithm is used to restrict the space defining the resulting image (Fig. 13a). The disadvantage of such an approach is that i) multiple features can be mapped to the same positions, and ii) the embedding can be relatively sparse, leaving a substantial amount of pixels unused. These issues were addressed in the REFINED approach [5] where the differences between distances in the feature space and 2D space were minimized using the Bayesian metric Multidimensional Scaling followed by a hill climbing approach. During the process, the positions of features were interchanged to minimize a cost function that measured the difference between the distances among the new feature locations to the estimated true distances (Fig. 13b). Most recently, a similar approach was taken in [100]. However, instead of minimizing the differences in distances, the goal was to minimize the difference between the ranking of pairwise distances between features and the ranking of pairwise distances between the coordinates of the assigned pixels. The minimization of the rankings was obtained using an iterative process. At each step, a pixel that had not been swapped for the longest time was identified and swapped with another pixel so that the difference between the rankings was minimized.
5 Discussion and open problems
In this section, we discuss several issues related to visualizations construction, such as the ability to compare different visualizations, the interpretability of visualizations and possible extensions of the proposed taxonomy. In the last subsection, we also elaborate on the connection between visualizations and their application in DCNN, establishing a future option to universally represent complex data for content-based retrieval.
5.1 Comparative evaluation of visualization approaches
We believe that one of the main issues with the reviewed papers is the lack of comparison between different visualization techniques. Quite often, we found a cluster of several articles that focused on a similar task, e.g., EEG or ECG classification. Although both the task and the underlying datasets are similar, different variants of visualizations are often proposed without sufficient discussion of their advantages over similar visualization approaches. Some exceptions to this statement exist, e.g., Zhu et al. [100] provided a thorough comparison of state-of-the-art techniques for unstructured visualizations with trained order. Overall, we believe that, in order to move this research domain forward, a battery of domain-specific evaluations is needed to reveal what visualization techniques are the most suitable for particular tasks and under what conditions.
In the DCNN compliance premises proposed in Sect. 3.1, we anticipated that there should be a great degree of smoothness in artificial visualizations. However, one can notice that some of the given examples perform well even with data that do not satisfy these premises, e.g., in malware detection. The main factor is the particular design of the DCNN, which is often ad-hoc or at least fine-tuned. If the original content does not have a great degree of smoothness and we perform pooling too early, we lose very important information. For example, for a binary executable file, the average of four consecutive bytes is meaningless. In this case, it is essential that the networks first find sufficiently suitable high-level concepts that will already have smoothness. This can be seen, for example, in \(\alpha {\text {Diff}}\), where the first two layers are convolutional with a high overlap of the convolutions. Therefore, compliance with individual premises is not always necessary. However, the employment of the mainstream pretrained networks, which are trained on photographs with a great degree of smoothness, must be revised in such cases.
On the other hand, many of the existing solutions are based on fully trained models. Given various kinds of visualizations, an opportunity arises to use these existing models for further transfer learning. If generated visualizations in another domain are similar to the original visualizations, it should be possible to fine-tune the model as in the above examples without requiring time-consuming and resource-intensive training from scratch.
5.2 Human interpretability
As an additional feature of the proposed visualization techniques, one can also consider the level of human understandability or interpretability. We perceive the concept of human interpretability as the ability of a domain expert to identify a particular object based on its visualization and/or ability to decode the features describing the object. To achieve this, the displayed entities, their positioning, and relations to other displayed entities should have an inherent meaning the community is familiar with, or such mapping must be easy to learn.
Human understandability is not a core requirement for a pipeline in which visualizations are consumed via DCNN or similar networks. Nonetheless, it may still provide the benefit of additional verification, explainability, or guidance. If a visualization is comprehensible for the domain expert, they may, e.g., point out possible errors or discrepancies between the object and its visualization, or suggest specific areas of interest within the visualization, which could guide or verify the classification models. As such, if two approaches provide end-results of similar quality, one should opt for the approach with more interpretable visualizations.
However, by its very nature, human interpretability is a somewhat noisy and subjective criterion. Some visualization techniques may be considered standard by a subset of the domain experts but be only vaguely known by others. Therefore, instead of a strict classification, we only provide a partial ordering of individual approaches, which may serve as pivots on the interpretability axis (see Fig. 14). We specifically focused on situations where similar visualization techniques have different interpretability based on some context features.
On the left side of Fig. 14, we depict several examples of supposedly highly interpretable visualizations. In [23], the authors used a standardized library to visualize chemical compounds (Fig. 14a). In [80], Chinese text is visualized sign by sign in natural order (Fig. 14b). In [52], one heartbeat is visualized in the same way that the ECG monitors display the electrical activity of the heart (adapted visualization in Fig. 14c). For all those cases, displayed entities and their position have a well-known interpretation. For example, in Fig. 14c, the X-axis denotes time, the Y-axis denotes heart electrical activity (e.g., current) and the line denotes particular values of the current in each time frame.
Compared to the previous cases, the central area of Fig. 14 contains examples that are only partially interpretable. Either some important knowledge needed for the interpretation is missing, or some non-trivial transformation of the source data was made (e.g., Fourier or wavelet transformations as in Fig. 14e). Hence, it may be challenging for humans to imagine the original data (i.e., perform an inverse transformation). Note, for example, the SuperTML approach [82], which is a derivative of the Super Character method [80] to visualize tabular data. While the original method (Fig. 14b) visualizes all necessary information for object identification (i.e., text), its version for tabular data (Fig. 14g) only depicts (re-ordered) feature values and discards corresponding feature labels. This makes object identification and feature mapping rather challenging. Similarly, for the gait recognition visualization (Fig. 14f), the ordering of skeleton joints is not depicted and must be remembered by the domain expert. Furthermore, there are numerous ways to map 3D space to RGB space, and a domain expert needs to have knowledge of the particular mapping used in the visualization.Footnote 7
In the last part of Fig. 14, we provide examples of visualizations that are not interpretable without a substantial learning effort of the domain expert. One general example is unstructured visualizations with trained order (see Fig. 14h), where feature values are usually normalized before visualization and their positions are trained for each dataset. Therefore, neither the color nor the position can provide a straightforward cue of what feature is represented on a specific spot.Footnote 8 This category would also include visualizations, where utilized data transformations are so complex that humans cannot recover the original data (i.e., performing an inverse transformation is virtually impossible for humans). One such example (see Fig. 14i) would be a modification of the base image through kernel multiplication, where the kernel represents object features [9].
5.3 Possible taxonomy extensions
We would like to emphasize that construction of the proposed visual taxonomy was case-driven, i.e., based on the reviewed papers. Some additional nodes or other taxonomy extensions are possible in the future.
One of the interesting directions that has not received much attention in the current context is the application of generative adversarial networks (GANs) [62]. In simple terms, GANs are trained to generate artificial visualizations that mimic the real data points from the distribution of training examples. The generation process is parameterized by a seed vector, and several applications showed that gradual modification of the seed vector resulted in a gradual modification of the output image [92]. However, note that feature entanglement is a common problem and therefore we cannot be sure that, e.g., the manipulation of a single seed dimension will have the same effect throughout its domain. While there are approaches trying to mitigate this issue [36, 92], the problem is currently only partially solved at best.
A straightforward way to use GANs in the current scenario is to apply a pretrained model (on an arbitrary domain), transform objects’ features into the seed vectors, and use GAN to generate images from the respective seed vectors. Such images may convey the similarity in a more natural way for subsequent DCNN models. Nevertheless, the feature entanglement may cause unexpected similarity fluctuations throughout the objects’ feature space. This may prove to be a limiting factor for this research direction. In the literature survey, we only found one example of GAN-based visualizations [79] utilizing the basic pipeline without much discussion of the potential limitations of this approach. Therefore, we believe this area is currently seriously under-explored and may foster some highly innovative research in the future.
From the taxonomy point of view, GAN-based visualizations can be considered as a special case of semi-structured visualizations as discussed in Sect. 4.2. However, the approach is sufficiently different from all other reviewed visualization techniques that it may deserve a special category in the future.
5.4 Toward universal feature extraction for complex data
As motivated in the introduction, an automated creation of suitable data visualizations constitutes the critical component of a framework / pipeline heading toward DCNN-based universal feature extractor for complex data [76]. In the reviewed cases, the visualizations were used with DCNN mostly for specific narrow tasks (such as classification) that seemingly had little in common as the tasks were tailored to specific domains. However, it was shown many times, that many deep features recognized in DCNN models are domain-agnostic, especially those at earlier vision layers (e.g., the pioneering study [19] or [56]). Hence, the shared part of all the reviewed cases might be found in the DCNN-based representation of the visualized data—the deep-feature vectors formed by neuron activations at a given DCNN layer. Detailed identification of such common features in the individual cases is the subject of our future work. The deep-feature vectors could be detached from the original narrow tasks and used as self-contained representations of the input data also in other tasks, either in general content-based retrieval, analytics or different specific tasks. As the feature vectors are technically high-dimensional semantic embeddings and share the same geometric universe (Euclidean vector space), they could be easily used with similarity search engines for effective and efficient content-based retrieval [99]. Thus, such an automated pipeline could lead to universal data representations for content-based retrieval in the future.
6 Conclusions
In this paper, we have reviewed domain cases where data visualizations were used with DCNNs for various domain-specific classification or retrieval tasks. Based on the review, we synthesized a taxonomy that provided a systematic overview of visualization techniques used with DCNN models. The taxonomy aims to enable the possibility of future systematic simplification of the domain-agnostic visualization design, leading to automation of the data processing pipeline that includes DCNN-based feature extraction or classification.
Notes
Note that on an intrinsic level, tabular data variables may be linked with complex relationships / hierarchical organization. However, such relationships are not captured in the data format.
Note that similarly as in tabular data, individual features of multivariate sequential data may or may not bear some intrinsic ordering of hierarchy, but these are not captured in the data format.
For example, datasets with added feature relationships (graphs, sequential data) can be transformed to tabular data simply by removing the information about feature relationships. Subsequently, visualization approaches suitable for tabular data can be applied.
In principle, other shapes are plausible, yet we did not found approaches aiming to display, e.g., circles. The main distinction is that the shape itself does not have an additional meaning (e.g., circles denoting one feature while squares another).
This type of visualization is mostly utilized for tabular data, where object features lack any explicit ordering or hierarchy.
As opposed to using individual features where the neighborhood information bears no meaning.
And also, they have to be able to imagine what kind of movement is represented, e.g., by a fast transition from greenish to yellowish color.
Note that with this respect, unstructured visualizations with arbitrary order may provide slightly more interpretable results as the position in visualization usually corresponds to the position in the source data.
References
Aakur SN, Narayanan S, Indla V et al (2021) Mg-Net: Leveraging pseudo-imaging for multi-modal metagenome analysis. In: de Bruijne M, Cattin PC, Cotin S et al (eds) Medical image computing and computer assisted intervention—MICCAI 2021. Springer International Publishing, Cham, pp 592–602
Ahn JW, Plaisant C, Shneiderman B (2013) A task taxonomy for network evolution analysis. IEEE Trans Vis Comput Graph 20(3):365–376
Al Rahhal MM, Bazi Y, Al Zuair M et al (2018) Convolutional neural networks for electrocardiogram classification. J Med Biol Eng 38(6):1014–1025. https://doi.org/10.1007/s40846-018-0389-7
Alvi RH, Rahman H, Khan AAS et al (2021) Deep learning approach on tabular data to predict early-onset neonatal sepsis. J Inf Telecommun 5(2):226–246. https://doi.org/10.1080/24751839.2020.1843121
Bazgir O, Zhang R, Dhruba SR et al (2020) Representation of features as images with neighborhood dependencies for compatibility with convolutional neural networks. Nat Commun. https://doi.org/10.1038/s41467-020-18197-y
Bozkir AS, Tahillioglu E, Aydos M et al (2021) Catch them alive: a malware detection approach through memory forensics, manifold learning and computer vision. Comput Secur 103(102):166. https://doi.org/10.1016/j.cose.2020.102166
Bromley J, Bentz J, Bottou L et al (1993) Signature verification using a Siamese time delay neural network. Int J Pattern Recognit Artif Intell 7:25. https://doi.org/10.1142/S0218001493000339
Brown T, Mann B, Ryder N et al (2020) Language models are few-shot learners. Adv Neural Inf Process Syst 33:1877–1901
Buturović L, Miljković D (2020) A novel method for classification of tabular data using convolutional neural networks. bioRxiv https://doi.org/10.1101/2020.05.02.074203,https://arxiv.org/abs/https://www.biorxiv.org/content/early/2020/05/03/2020.05.02.074203.full.pdf
Chen J, Hu K, Yu Y et al (2020) Software visualization and deep transfer learning for effective software defect prediction. In: Proceedings of the ACM/IEEE 42nd international conference on software engineering. association for computing machinery, New York, NY, USA, ICSE ’20, p 578-589, https://doi.org/10.1145/3377811.3380389
Chengzhi Q, Chenghu Z, Tao P (2003) Taxonomy of visualization techniques and systems–concerns between users and developers are different. In: Asia GIS Conference, p 37
Chi EHH (2000) A taxonomy of visualization techniques using the data state reference model. In: IEEE symposium on information visualization 2000. INFOVIS 2000. Proceedings, IEEE, pp 69–75
Chiu CH, Tsai YC (2020) Use text mining for financial reports analysis: long text to image converter. In: 2020 the 6th international conference on communication and information processing. Association for computing machinery, New York, NY, USA, ICCIP 2020, pp 7–11, https://doi.org/10.1145/3442555.3442557
Cui Z, Xue F, Cai X et al (2018) Detection of malicious code variants based on deep learning. IEEE Trans Ind Inf 14(7):3187–3196. https://doi.org/10.1109/TII.2018.2822680
Dastile X, Celik T (2021) Making deep learning-based predictions for credit scoring explainable. IEEE Access 9:50,426-50,440. https://doi.org/10.1109/ACCESS.2021.3068854
Debayle J, Hatami N, Gavet Y (2018) Classification of time-series images using deep convolutional neural networks. In: Tenth international conference on machine vision (ICMV 2017), SPIE, pp 242–249, https://doi.org/10.1117/12.2309486
Deng J, Dong W, Socher R et al (2009) ImageNet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, IEEE, pp 248–255
Devlin J, Chang MW, Lee K et al (2018) BERT: pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805
Donahue J, Jia Y, Vinyals O et al (2014) Decaf: a deep convolutional activation feature for generic visual recognition. In: Proceedings of the 31st international conference on international conference on machine learning, vol 32. JMLR.org, ICML’14, p I–647–I–655
Dosovitskiy A, Beyer L, Kolesnikov A et al (2010) An image is worth 16x16 words: transformers for image recognition at scale. arxiv 2020. arXiv preprint arXiv:2010.11929
Elias P, Sedmidubsky J, Zezula P (2015) Motion images: an effective representation of motion capture data for similarity search. In: Amato G, Connor R, Falchi F et al (eds) Similarity search and applications. Springer International Publishing, Cham, pp 250–255
Feng G, Li B, Yang M et al (2018) V-CNN: data visualizing based convolutional neural network. In: 2018 IEEE international conference on signal processing, communications and computing (ICSPCC), pp 1–6. https://doi.org/10.1109/ICSPCC.2018.8567781
Fernandez M, Ban F, Woo G et al (2018) Toxic colors: the use of deep learning for predicting toxicity of compounds merely from their graphic images. J Chem Inf Model 58(8):1533–1543. https://doi.org/10.1021/acs.jcim.8b00338
Fischer P, Dosovitskiy A, Brox T (2014) Descriptor matching with convolutional neural networks: a comparison to sift. CoRR arXiv:1405.5769
Gao Y, Zhang Y, Wang H et al (2019) Decoding behavior tasks from brain activity using deep transfer learning. IEEE Access 7:43,222-43,232. https://doi.org/10.1109/ACCESS.2019.2907040
Gao Y, Gao B, Chen Q et al (2020) Deep convolutional neural network-based epileptic electroencephalogram (EEG) signal classification. Front Neurol. https://doi.org/10.3389/fneur.2020.00375
Georgiou T, Liu Y, Chen W et al (2020) A survey of traditional and deep learning-based feature descriptors for high dimensional data in computer vision. Int J Multim Inf Retr 9(3):135–170. https://doi.org/10.1007/s13735-019-00183-w
Gleicher M, Albers D, Walker R et al (2011) Visual comparison for information visualization. Inf Vis 10(4):289–309
Goh GB, Siegel C, Vishnu A et al (2017) Chemception: a deep neural network with minimal chemistry knowledge matches the performance of expert-developed QSAR/QSPR models. arxiv:1706.06689
He K, Zhang X, Ren S et al (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hetland ML, Skopal T, Lokoc J et al (2013) Ptolemaic access methods: Challenging the reign of the metric space model. Inf Syst 38(7):989–1006. https://doi.org/10.1016/j.is.2012.05.011
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Huerta Á, Martínez-Rodrigo A, Puchol A et al (2020) Comparison of pre-trained deep learning algorithms for quality assessment of electrocardiographic recordings. In: 2020 international conference on e-health and bioengineering (EHB), pp 1–4, https://doi.org/10.1109/EHB50910.2020.9280217
Jain V, Goel M, Shah K (2022) Deep learning on small tabular dataset: Using transfer learning and image classification. In: Dev A, Agrawal SS, Sharma A (eds) Artificial Intelligence and Speech Technology. Springer International Publishing, Cham, pp 555–568
Karayev S, Trentacoste M, Han H et al (2014) Recognizing image style. In: Proceedings of the British machine vision conference. BMVA Press, https://doi.org/10.5244/C.28.122
Karras T, Laine S, Aittala M et al (2020) Analyzing and improving the image quality of stylegan. In: 2020 IEEE/CVF conference on computer vision and pattern recognition, CVPR 2020, Seattle, WA, USA, June 13-19, 2020. IEEE, pp 8107–8116, https://doi.org/10.1109/CVPR42600.2020.00813
Keim DA (2002) Information visualization and visual data mining. IEEE Trans Vis Comput Graph 8(1):1–8
Keim DA, Kriegel HP (1996) Visualization techniques for mining large databases: a comparison. IEEE Trans Knowl Data Eng 8(6):923–938
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, vol 25. Curran Associates, Inc., p 1097–1105
Kuang B, Nnabuife SG, Rana Z (2021) Pseudo-image-feature-based identification benchmark for multi-phase flow regimes. Chem Eng J Adv 5(100):060. https://doi.org/10.1016/j.ceja.2020.100060
Lad S, Adamuthe A (2020) Malware classification with improved convolutional neural network model. Int J Comput Netw Inf Secur 12:30–43. https://doi.org/10.5815/ijcnis.2020.06.03
Lee B, Plaisant C, Parr CS et al (1967/1983) Semiology of graphics: diagrams, networks, maps. ACM Press, New York
Lee B, Plaisant C, Parr CS et al (2006) Task taxonomy for graph visualization. In: Proceedings of the 2006 AVI workshop on BEyond time and errors novel evaluation methods for information visualization—BELIV ’06. ACM Press, https://doi.org/10.1145/1168149.1168168
Liu B, Huo W, Zhang C et al (2018) \(\alpha \)Diff: cross-version binary code similarity detection with DNN. Association for Computing Machinery, New York, pp 667–678. https://doi.org/10.1145/3238147.3238199
Lohse GL, Biolsi K, Walker N et al (1994) A classification of visual representations. Commun ACM 37(12):36–50
Ma S, Zhang Z (2018) OmicsMapNet: transforming omics data to take advantage of deep convolutional neural network for discovery. arxiv:1804.05283
Van der Maaten L, Hinton G (2008) Visualizing data using t-SNE. J Mach Learn Res 9(11):2579–2605
Mackinlay J (1986) Automating the design of graphical presentations of relational information. ACM Trans Graph (Tog) 5(2):110–141
Mallik A, Khetarpal A, Kumar S (2022) ConRec: malware classification using convolutional recurrence. J Comput Virol Hacking Tech. https://doi.org/10.1007/s11416-022-00416-3
Mayampurath A, Sanchez-Pinto LN, Carey KA et al (2019) Combining patient visual timelines with deep learning to predict mortality. PLOS ONE 14(7):1–12. https://doi.org/10.1371/journal.pone.0220640
Mohd Shaid SZ, Maarof M (2014) Malware behaviour visualization. Jurnal Teknologi. https://doi.org/10.11113/jt.v70.3512
Mohebbanaaz Kumar LVR, Sai YP (2022) A new transfer learning approach to detect cardiac arrhythmia from ECG signals. Signal Image Video Process. https://doi.org/10.1007/s11760-022-02155-w
Nahak S, Pathak A, Saha G (2023) Evaluation of handcrafted features and learned representations for the classification of arrhythmia and congestive heart failure in ECG. Biomed Signal Process Control 79(104):230. https://doi.org/10.1016/j.bspc.2022.104230
Nataraj L, Karthikeyan S, Jacob G et al (2011) Malware images: visualization and automatic classification. In: International symposium on visualization for cyber security (VizSec), https://vision.ece.ucsb.edu/sites/default/files/publications/nataraj_vizsec_2011_paper.pdf
Naz M, Shah JH, Khan MA et al (2021) From ECG signals to images: a transformation based approach for deep learning. PeerJ Comput sci 7:e386–e386. https://doi.org/10.7717/peerj-cs.386
Neyshabur B, Bhojanapalli S, McAllester D et al (2017) Exploring generalization in deep learning. In: Advances in neural information processing systems, vol 30
O’Kane TW, Fife D (2021) Multivariate visualization: a taxonomy and novel visualization methods. PsyArXiv
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22(10):1345–1359
Pfitzner D, Hobbs V, Powers D (2003) A unified taxonomic framework for information visualization. In: Proceedings of the Asia-pacific symposium on information visualisation, vol 24. pp 57–66
Pinto JP, Pimenta A, Novais P (2021) Deep learning and multivariate time series for cheat detection in video games. Mach Learn 110(11):3037–3057. https://doi.org/10.1007/s10994-021-06055-x
Radford A, Kim JW, Hallacy C et al (2021) Learning transferable visual models from natural language supervision. CoRR arXiv:2103.00020
Reed S, Akata Z, Yan X et al (2016) Generative adversarial text to image synthesis. In: Balcan MF, Weinberger KQ (eds) Proceedings of The 33rd international conference on machine learning, proceedings of machine learning research, vol 48. PMLR, New York, pp 1060–1069, https://proceedings.mlr.press/v48/reed16.html
Ren Z, Chen G, Lu W (2020) Malware visualization methods based on deep convolution neural networks. Multimed Tools Appl 79(15):10,975-10,993. https://doi.org/10.1007/s11042-019-08310-9
Rifaioglu AS, Nalbat E, Atalay V et al (2020) DEEPScreen: high performance drug–target interaction prediction with convolutional neural networks using 2-d structural compound representations. Chem Sci 11(9):2531–2557. https://doi.org/10.1039/c9sc03414e
Rodrigues JF Jr, Traina AJ, de Oliveira MCF et al (2007) The spatial-perceptual design space: a new comprehension for data visualization. Inf Vis 6(4):261–279
Salem M, Taheri S, Yuan J (2018) ECG arrhythmia classification using transfer learning from 2- dimensional deep CNN features. In: 2018 IEEE biomedical circuits and systems conference (BioCAS), pp 1–4. https://doi.org/10.1109/BIOCAS.2018.8584808
Samek W, Montavon G, Vedaldi A et al (2019) Explainable AI: interpreting, explaining and visualizing deep learning. Springer, Berlin
Scheibel W, Trapp M, Limberger D et al (2020) A taxonomy of treemap visualization techniques. In: VISIGRAPP (3: IVAPP), pp 273–280
Schulz HJ, Nocke T, Heitzler M et al (2013) A design space of visualization tasks. IEEE Trans Vis Comput Graph 19(12):2366–2375
Sharma A, Vans E, Shigemizu D et al (2019) DeepInsight: a methodology to transform a non-image data to an image for convolution neural network architecture. Sci Rep. https://doi.org/10.1038/s41598-019-47765-6
Shneiderman B (1992) Tree visualization with tree-maps. ACM Trans Graph 11(1):92–99. https://doi.org/10.1145/102377.115768
Shneiderman B (1996) The eyes have it: a task by data type taxonomy for information visualizations. In: Proceedings 1996 IEEE symposium on visual languages. IEEE Comput. Soc. Press, https://doi.org/10.1109/vl.1996.545307
Silva P, Luz E, Silva G et al (2020) Towards better heartbeat segmentation with deep learning classification. Sci Rep 10(1):20,701. https://doi.org/10.1038/s41598-020-77745-0
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Skopal T (2007) Unified framework for fast exact and approximate search in dissimilarity spaces. ACM Trans Database Syst 32(4):29. https://doi.org/10.1145/1292609.1292619
Skopal T (2020) On visualizations in the role of universal data representation. In: Gurrin C, Jónsson BÞ, Kando N et al (eds) Proceedings of the 2020 on international conference on multimedia retrieval, ICMR 2020, Dublin, Ireland, June 8-11, 2020. ACM, pp 362–367, https://doi.org/10.1145/3372278.3390743
Skopal T, Bustos B (2011) On nonmetric similarity search problems in complex domains. ACM Comput Surv 43(4):34:1-34:50. https://doi.org/10.1145/1978802.1978813
Sotiroudis SP, Sarigiannidis P, Goudos SK et al (2021) Fusing diverse input modalities for path loss prediction: a deep learning approach. IEEE Access 9:30,441-30,451. https://doi.org/10.1109/ACCESS.2021.3059589
Sreehari S (2020) Visualization transforms of non-spatial data for convolutional neural networks. In: Zelinski ME, Taha TM, Howe J et al (eds) Applications of Machine Learning 2020, International Society for Optics and Photonics, vol 11511. SPIE, Bellingham, pp 83–91. https://doi.org/10.1117/12.2572485
Sun B, Yang L, Dong P et al (2018) Super characters: a conversion from sentiment classification to image classification. In: Proceedings of the 9th workshop on computational approaches to subjectivity, sentiment and social media analysis. Association for computational linguistics, Brussels, Belgium, pp 309–315, https://doi.org/10.18653/v1/W18-6245, https://aclanthology.org/W18-6245
Sun B, Yang L, Chi C et al (2019a) Squared english word: a method of generating glyph to use super characters for sentiment analysis. In: Chhaya N, Jaidka K, Sinha AR et al (eds) Proceedings of the 2nd workshop on affective content analysis (AffCon 2019) co-located with thirty-third AAAI conference on artificial intelligence (AAAI 2019), Honolulu, USA, January 27, 2019, CEUR Workshop Proceedings, vol 2328. CEUR-WS.org, pp 140–151, http://ceur-ws.org/Vol-2328/4_2_paper_21.pdf
Sun B, Yang L, Zhang W et al (2019b) SuperTML: two-dimensional word embedding for the precognition on structured tabular data. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (CVPR) workshops
Szegedy C, Liu W, Jia Y et al (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Tan M, Le QV (2019) Efficientnet: rethinking model scaling for convolutional neural networks. In: Chaudhuri K, Salakhutdinov R (eds) Proceedings of the 36th international conference on machine learning, ICML 2019, 9–15 June 2019, Long Beach, California, USA, Proceedings of Machine Learning Research, vol 97. PMLR, pp 6105–6114, http://proceedings.mlr.press/v97/tan19a.html
Tang B, Wang Y (2012) Design of large-scale sensory data processing system based on cloud computing. Res J Appl Sci Eng Technol 4(8):1004–1009
Tory M, Moller T (2004) Rethinking visualization: a high-level taxonomy. In: IEEE symposium on information visualization, IEEE, pp 151–158
Tweedie L (1997) Characterizing interactive externalizations. In: Proceedings of the ACM SIGCHI conference on human factors in computing systems, pp 375–382
Vasan D, Alazab M, Wassan S et al (2020) IMCFN: image-based malware classification using fine-tuned convolutional neural network architecture. Comput Netw 171(107):138. https://doi.org/10.1016/j.comnet.2020.107138
Vu DL, Nguyen TK, Nguyen TV et al (2019) A convolutional transformation network for malware classification. In: 2019 6th NAFOSTED conference on information and computer science (NICS), pp 234–239. https://doi.org/10.1109/NICS48868.2019.9023876
Várkonyi DT, Seixas JL, Horváth T (2023) Dynamic noise filtering for multi-class classification of beehive audio data. Expert Syst Appl 213(118):850. https://doi.org/10.1016/j.eswa.2022.118850
Wan J, Wang D, Hoi SCH et al (2014) Deep learning for content-based image retrieval: a comprehensive study. In: MM ’14. ACM, pp 157–166
Wu Z, Lischinski D, Shechtman E (2021) Stylespace analysis: disentangled controls for stylegan image generation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 12863–12872
Xu G, Shen X, Chen S et al (2019) A deep transfer convolutional neural network framework for EEG signal classification. IEEE Access 7:112767–112776. https://doi.org/10.1109/ACCESS.2019.2930958
Xu Y, Liu Z, Li Y et al (2020) Feature data processing: making medical data fit deep neural networks. Fut Gener Comput Syst 109:149–157. https://doi.org/10.1016/j.future.2020.02.034
Xu ZW, Liu XM, Zhang K (2019) Mechanical properties prediction for hot rolled alloy steel using convolutional neural network. IEEE Access 7:47,068-47,078. https://doi.org/10.1109/ACCESS.2019.2909586
Yang CL, Chen ZX, Yang CY (2020) Sensor classification using convolutional neural network by encoding multivariate time series as two-dimensional colored images. Sensors. https://doi.org/10.3390/s20010168
Yue S, Wang T (2017) Imbalanced malware images classification: a CNN based approach. arXiv:1708.08042
Zeiler MD, Fergus R (2017) Visualizing and understanding convolutional networks. CoRR arXiv:1311.2901
Zezula P, Amato G, Dohnal V et al (2005) Similarity search: the metric space approach (Advances in database systems). Springer-Verlag, Berlin, Heidelberg
Zhu Y, Brettin T, Xia F et al (2021) Converting tabular data into images for deep learning with convolutional neural networks. Sci Rep. https://doi.org/10.1038/s41598-021-90923-y
Funding
This work was supported by the Czech Science Foundation (GAČR), grant number 22-21696 S.
Author information
Authors and Affiliations
Contributions
TS derived the original idea behind the manuscript and wrote the introduction and motivation parts. LP, DH, IS and DB contributed to the review by searching, reading and classifying related papers. All authors contributed to writing the main review text. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Skopal, T., Peška, L., Hoksza, D. et al. Visualizations for universal deep-feature representations: survey and taxonomy. Knowl Inf Syst 66, 811–840 (2024). https://doi.org/10.1007/s10115-023-01933-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-023-01933-3