Abstract
Prediction and classification of diseases are essential in medical science, as it attempts to immune the spread of the disease and discover the infected regions from the early stages. Machine learning (ML) approaches are commonly used for predicting and classifying diseases that are precisely utilized as an efficient tool for doctors and specialists. This paper proposes a prediction framework based on ML approaches to predict Hepatitis C Virus among healthcare workers in Egypt. We utilized real-world data from the National Liver Institute, founded at Menoufiya University (Menoufiya, Egypt). The collected dataset consists of 859 patients with 12 different features. To ensure the robustness and reliability of the proposed framework, we performed two scenarios: the first without feature selection and the second after the features are selected based on sequential forward selection (SFS). Furthermore, the feature subset selected based on the generated features from SFS is evaluated. Naïve Bayes, random forest (RF), K-nearest neighbor, and logistic regression are utilized as induction algorithms and classifiers for model evaluation. Then, the effect of parameter tuning on learning techniques is measured. The experimental results indicated that the proposed framework achieved higher accuracies after SFS selection than without feature selection. Moreover, the RF classifier achieved 94.06% accuracy with a minimum learning elapsed time of 0.54 s. Finally, after adjusting the hyperparameter values of the RF classifier, the classification accuracy is improved to 94.88% using only four features.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Hepatitis C Virus (HCV) is a disease affecting the human population on a global level. It is a blood-borne infection that can spread through direct contact with an infected person’s blood or body fluids containing blood. According to the World Health Organization (WHO), hepatitis C is a global disease. The WHO estimated that about 58 million people have chronic HCV infection, with about 1.5 million new infections occurring annually [1]. Poor developing countries of Asia and Africa show the highest prevalence of this infection compared to developed countries in Europe and North America. Furthermore, in countries like Pakistan, China, and Egypt, the number of people with chronic diseases is higher [2, 3].
There needs to be more information about the prevalence of HCV among Egyptian healthcare workers (HCWs). HCWs are frequently exposed to different biological agents during activities and are frequently monitored. So, healthcare professionals who closely interact with patients in Egypt are at increased risk of HCV infection and other blood-borne pathogens [4].
Hence, we badly need accurate and reliable noninvasive technology to diagnose HCV. Machine learning (ML) algorithms are particularly adept at analyzing medical phenomena by capturing complex and nonlinear relationships in clinical data. The ML algorithms, such as classification techniques, can be utilized to develop a model to diagnose HCV by identifying people who have been infected with the virus. However, inappropriate characteristics in the attribute set can spoil the classifier's performance [5]. Feature selection defines a subset of features or variables that describe data to obtain a more compact and essential representation of the available information and ignore all other redundant and irrelevant features [6]. Feature selection is a powerful way to enhance the functioning and reduce the model development time of a classifier.
Ensemble learning is a generic meta-machine learning technique that attempts to improve predictive performance by mixing predictions from many models. It is further a flexible algorithm that predicts using supervised learning following training with numerous individual models or weak learners. There are several ensemble strategies available, with bagging and boosting being the most well known. Random forest and extra tree algorithm are common bagging methods, whereas gradient boosting, adaboost, and extreme gradient boosting are popular boosting methods. In theory, ensembles are hypothetical to produce more effective results if the models are varied in a significant way, even though some normal decision tree algorithms can produce better ensembles than deliberate ones. Although it may create an apparently infinite number of ensembles for any predictive modeling challenge, three strategies dominate the field of ensemble learning. The three primary classes of ensemble learning methods are bagging, stacking, and boosting, and it is critical to grasp each approach and include them in the predictive mode. Bagging includes fitting many decision trees on various instances of the dataset and averaging the predictions. Stacking involves fitting many different model types on the same data and using another model to learn how to best combine the predictions. Boosting involves adding ensemble members sequentially that correct the predictions made by prior models and outputs a weighted average of the predictions [7, 8]. In this work, the best score is obtained through a kind of bagging ensemble approach which is random forest (RF).
2 Research questions (RQ)
The research question of this study includes the following points.
-
How effective is the proposed model in assisting Hepatitis C Virus (HCV) among healthcare workers (HCWs) in Egypt?
-
What influence do the proposed ensemble learning models have on classification results, and how might they address the overfitting issue?
-
Why was the SFS feature selection method used instead of the gain ratio and genetic algorithm and how is the feature selection useful for the data?
3 Research objectives
The main objective of this paper is to build an ML framework (MHF_HCV) for diagnosing HCV disease among HCWs in Egypt. The MHF_HCV framework used the classification and feature selection approach on a real-world HCV dataset to predict HCV. First, we examined the accuracy of well-known ML algorithms in classifying and diagnosing HCV. After that, we are introduced and implemented a hybrid prediction model using sequential forward selection (SFS)-based wrapper feature selection and classification. Then, the effect of parameter tuning on learning techniques is measured. Finally, the performance of different classifiers is compared in terms of accuracy, recall, precision, and F1 scores. The primary motivation and objectives of this study can be summarized as follows:
-
We executed two scenarios, the first without feature selection and the second with feature selection based on Sequential Forward Selection (SFS), to test the robustness and reliability of the proposed framework.
-
Additionally, an evaluation of the feature subset chosen based on the SFS-generated features is carried out. As an induction technique and classifiers for model evaluation, Naive Bayes (NB), random forest (RF), K-nearest neighbor (KNN), and logistic regression (LR) are used.
-
The impact of parameter tuning on learning methods is then evaluated. According to the experiment results, the proposed framework had better accuracy with SFS selection than those without.
-
Finally, with only four features, the classification accuracy is enhanced to 94.88% after modifying the RF classifier's hyperparameter values.
4 Research contributions
The main contribution of this work is summarized as follows:
-
A practical model is designed to predict HCV disease in HCV patients and HCWs in Egypt by choosing the essential features that improve the classification of HCWs in Egypt if they have HCV disease.
-
The performance of well-known classifiers is evaluated using all features from a real-world HCV dataset for Egyptian healthcare workers.
-
The best attributes are selected using SFS-based wrapper feature selection algorithm, which selects features that maximize classification accuracy.
-
The performance of classifiers is measured on all features and selected features from the real-world HCV dataset.
-
The classifier’s performance is enhanced by fine-tuning the hyper-parameters, which also affects the quality of features selected using the SFS method wrapped with the classifier.
-
The best combination of feature set and classifier along with its tuned hyper-parameters is returned for accurate classification HCV.
-
The proposed method can be applied and integrated into real-life applications which can help experts in decision-making.
The rest of this paper is organized as follows: Sect. 2 presents the motivation and problem formulation, while the detailed discussion of the related work is investigated in Sect. 3. Section 4 explains our proposed method, and Sect. 5 demonstrates the experimental results. Section 6 presents a discussion of the results. Section 7 presents the limitation and threads of the current study. Finally, Sect. 7 offers the conclusion and future work.
5 Motivation and problem formulation
HCWs represent a high-risk population for sharps injuries, needles, and scalpels during the execution of their health care duties. Therefore, HCWs are at risk of infection by caring for patients infected with HCV infections. This motivates academics and researchers to devise a framework that can help predict HCV disease at an early stage among HCWs.
The HCV dataset for HCWs collected from the National Liver Institute (NLI), founded at Menoufiya University (Menoufiya, Egypt) and used in the proposed work for developing the ML framework. The HCV dataset D, along with output class Y, consists of feature set X = {X1; X2; _ _ _; Xn} with n features and the instances J = {J1; J2; _ _ _; Jm} corresponding to m subjects (cases).
Definition 1
A dataset D is the composition of instances J = {Jk|1 ≤ k < m}, where m is the total number of subjects (cases) and feature set X = {Xi|1 ≤ i < n}, where n is the number of features.
Definition 2
An instance Jk is represented by feature values Xi, such as Jk = {Xi 1 ≤ i < n} and n is the number of features in D. The value Xi is either categorical or numeric.
The goal of designing a framework for accurate predictions (PR) using a learning algorithm (C) is to make the predictive model learn to fit by analyzing the data behavior and converge by reducing the error (ER) present in all instances (J) collectively as depicted in Eq. 1.
The prediction PR from C is evaluated based on performance (P), measured using metrics accuracy, precision, recall, and F1-scores. The metrics are explained in detail in Sect. 4.6.
6 Related work
The current studies prove that 20% of people with viral hepatitis “C” develop symptoms of influenza-like symptoms of the disease. In comparison, 80% of other people with the disease do not feel symptoms, but the infection remains [7, 8]. Therefore, it is essential to undergo frequent tests to ensure that the body is free of infection, mainly if the patient receives medications through needles, which help transmit the disease. Egypt recorded the highest prevalence of hepatitis C in the world, and this epidemic is expected to reach its peak soon [9]. According to a 2010 study, an estimated more than half a million people contract the virus for the first time each year. At the same time, the Egyptian Ministry of Health and Population forecasts that the number of cases of HIV infection annually is 100,000 people. Studies have shown that the infection rate with HCV in Egypt is the highest in the world, as it is ten times higher than in Europe and America [10,11,12,13]. A framework that integrates data mining with decision tree (DT) and fuzzy logic is presented by Ali et al. [16] to manage and predict HCV cases. They utilized the trapezoidal fuzzy number (TFN) that achieves 98.1% compared with 92.5% prediction results by DT. Abd El-Salam et al. performed a laboratory analysis study of 4962 HCV patients in Egypt between 2006 and 2017 based on ML [17]. They used 24 clinical laboratory variables, and the results investigated 2218 patients infected with Esophageal Varices and not present in 2744 patients. Their model used six well-known classifiers, including neural networks (NN), Naïve Bayes (NB), DT, support vector machine (SVM), RF, and Bayesian network (BN). They utilized data collected from the Egyptian National Committee to Combat Viral Hepatitis in the national treatment program for patients with viral hepatitis in Egypt under the supervision of the Ministry of Health. The accuracies obtained are 67.8%, 66.3%, 67.2%, 65.6%, 66.7%, and 68.9% using support vector machine (SVM), RF, C4.5, multi-layer perceptron (MLP), NB, and BN, respectively.
Nandipati et al. [18] present an HCV prediction model using ML approaches based on random forest (RF) and k-nearest neighbors algorithm (KNN) classifiers for the applied dataset (HCV) found in the UCI-ML repository [19]. They utilized 668 instances with mild to moderate class 0 and cirrhosis with class 1. They used Python and R for programming with different features and attributes. The combination of components can be utilized more efficiently based on ML to provide improved insights into antibody sequences that affect the HCV response [20]. They used ML approaches to predict the clinical groups that combine the features to identify the most significant features using RF classification.
Hashem et al. [21] present ML approaches to predict hepatocellular carcinoma with HCV-related chronic liver disease. They present a set of input variables that are filtered to get the optimal variable subset based on LR, DT, and classification and regression tree (CART). The accuracies abstained were 96%, 99%, and 95.5% using LR, DT, and CART, respectively.
The prediction of HCV virus results from viral nucleotides using several combinations of ML approaches is presented by KayvanJoo et al. [22]. They used DT, SVM, NB, and NN to predict the interferon-alpha (IFN-alpha) and ribavirin (RBV) therapy response based on processed features. They produce 10 attribute weighting models from 76 overall attributes for the initial dataset. These eleven attributes include Chi-square, Gini index, Deviation, Info-Gain, Info-Gain Ratio, SVM, PCA, Uncertainty, Relief, and Rule. The eleven attributes are then classified based on SVM, NB, NN, and DT, and the average accuracy is 85%. The summary of the most recent methodologies and efforts for predicting HCV cases is investigated in Table 1. The table refers to the authors, the utilized dataset, the number of instances or patients or collected issues, and the feature selections that refer to which the authors used feature selection.
Further, the authors used ML methodologies and the resulting performance metrics. The current efforts for another diagnosis of diseases are presented by Alade, et al. [23] by which a neural network model is presented to help diagnose pregnant women if they have diabetes or not. The used dataset was obtained from a medical database, the Pima Indian Database. Moreover, Abayomi-Alli et al. [24] developed a system to enhance the early detection of PD using a deep learning network called bidirectional LSTM (BiLSTM) through data augmentation for tiny datasets, while Ogundokun et al. [25] used two computational intelligence methods, which are decision tree (DT) and K-nearest neighbor (KNN) for heart disease detection. They utilized the autoencoder feature extraction algorithm to minimize the features required to describe the heart disease dataset. The heart disease databases from the National Health Service (NHS) database are used to evaluate the performance of each method. Furthermore, a fine-tuning algorithm based on fuzzy KNN applied to the pregnancies diabetes dataset called (PIMA) is presented by Salem et al. [26]. Ensemble learning is a powerful technique in machine learning that combines the predictions of multiple models to improve the overall performance. The idea behind ensemble learning is that by combining the predictions of multiple models, the ensemble can leverage the strengths of each individual model and reduce the weaknesses. Ensemble methods can often achieve better performance than individual models, as they can capture a broader range of patterns in the data.
-
Robustness Ensemble methods are more robust to overfitting than individual models. By combining multiple models, the ensemble can better handle noisy or uncertain data.
-
Diversity Ensemble methods can be used to create diverse sets of models, which can help to reduce the risk of overfitting and improve the overall performance.
-
Handling imbalance data Ensemble methods can handle imbalance data well by combining multiple models with different learning strategies.
-
Handling high-dimensional data Ensemble methods can be used to effectively reduce the dimensionality of high-dimensional datasets, which can improve the performance of the model.
-
Handling complex data Ensemble methods can handle complex data by combining multiple models with different architectures or learning algorithms.
In short, ensemble learning is a powerful technique that can improve the performance, robustness, and interpretability of machine learning models. Ensemble methods can be used to improve the performance of various tasks such as classification, regression, and anomaly detection and in various fields such as in sport science, agriculture, wireless sensor network [8, 27,28,29,30,31].
From the mentioned studies, we can conclude that the efforts made to predict viral hepatitis, especially in Egypt, are the most interesting and required to address the spread of the virus. However, the related work focused only on patients in general and not dealing with hospital staff in mind. Moreover, the accuracy based on known traditional classifiers must be improved. Therefore, the diversity of the database used and the number of instances generated are not a fair comparison between the results obtained. Moreover, most studies mentioned above used a well-known dataset rather than a real-world datasets. Also, the earlier studies were only concerned with studying the effectiveness of hyperparameter tuning on the classification performance. Therefore, in this study, we used a data set collected from actual working hospital patients to predict HBV infection incidence from registered cases. We also identified the most relevant features using the SFS feature selection algorithm that achieved accuracy compared to the accuracy obtained before feature selection. Moreover, studying the hyperparameter tuning effects can enhance the quality of features selected using the SFS method, which is dependent on the classifier performance.
7 Methodology
The proposed system, MHF_HCV, has been designed to classify patients infected with HCV from those who are not. We worked on improving the accuracy of ML classification methods for the prognosis of HCV among HCWs in Egypt. The performance of the classifiers has been tested on all attributes and selected features separately to compare the achieved accuracy. To identify the important features and improve the efficiency of the classification process, an SFS-based wrapper feature selection approach was used to find the optimal feature subset that influences the class discovery process and improves the classification accuracy. Popular ML classifiers were used on these to classify selected features into different classes. The methodology of the proposed system is structured into six stages which include: (1) data gathering, (2) data pre-processing; (3) data splitting; (4) feature selection; (5) ML classifiers; (6) classifier performance evaluation. Figure 1 shows the framework for predicting HCV disease.
7.1 Description of the dataset
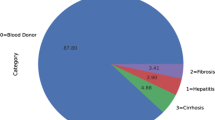
In this study, we used a real-world dataset of hepatitis C prevalence among HCWs in Egypt, with the highest prevalence of HCV [2]. This dataset was obtained from The NLI institute, founded at Menoufiya University (Menoufiya, Egypt). The dataset consists of 859 records (patients) with 12 features, which present test information of each patient. In addition, 11 attributes of these features are taken as diagnosis inputs, whereas the ‘HCV_PCR’ attribute is selected as output. Table 2 briefly describes the HCV dataset used in our study.
7.2 Data preprocessing
Data preprocessing is the first step in the proposed system to remove the noisy value and replace the missing values of some attributes. The dataset for experiments has some missing records; it is assumed that missing, inconsistent, and duplicate data have been resolved. Therefore, the missing records are removed from the data set, and most of the medical attributes used were transformed from numerical to categorical data, as shown in Table 2.
7.3 Data splitting
This section explains different splitting algorithms used in our study.
7.3.1 K-fold cross-validation
A cross-validation algorithm is a method commonly used in ML. The primary purpose of cross-validation is to achieve a stable and confident estimate of the model performance. In K-fold cross-validation, data are split into k different parts. For each iteration, k − 1 parts are used to train the model, and the remaining part is used as a validation set. The process is iterated according to the number of folds. The model’s generalization performance is the average of the estimated scores [32].
7.3.2 Train–test split
Train–test splits the dataset into random train and test subsets. This method depends on the size of the dataset [33].
7.4 Feature selection
The primary purpose of the feature selection process is to select a subset of features from the original features that maximize classification accuracy [34]. In this study, an SFS algorithm based on the wrapper selection approach was proposed for feature selection to select the best subset of features due to its simplicity and empirically successful. The wrapper model approach uses the classifier as an induction algorithm to measure the good features subset [35]. Wrappers typically perform better in choosing features since they employ a predetermined classification algorithm rather than an independent measure for the subset evaluation. Additionally, it considers the interactions between features with less computation. Wrapper methods generally achieve a better accuracy rate and use cross-validation to avoid over-fitting. Here, tenfold cross-validation is used on the training set for classification to evaluate the selected feature. SFS is the most straightforward greedy search algorithm [36]. It is a bottom-up search method that starts with an empty set of features and sequentially adds features selected based on some evaluation function, reducing the mean square error rate.
7.5 Classification process
Classification algorithms used to assign a class for an unseen record accurately are utilized in this study. Moreover, well-known classifiers NB, RF, KNN, and LR are applied to investigate the contributions of the features selected by the SFS method to the classification accuracy.
7.5.1 Naïve Bayes (NB)
NB [37, 38] is a widely used method for classification and is particularly suitable when the input dimensionality is high. Despite its simplicity, NB can often outperform more complex classification techniques. This is because it measures the probability of each input feature (attribute) for a predictable state.
The Bayesian classifier uses the Bayes rule to calculate the posterior probability for each class ci.
where
where P (Ci): the Apriori likelihood of class Ci. P(y): the likelihood density for feature y. P (y|Ci): the class conditional likelihood density of the feature y that belongs to the Ci class. P (Ci|y): the posterior probability of the Ci class when observing y.
7.5.2 Random forests (RF)
The RF [39] is defined as an ensemble learning method for classification and regression. Ensemble learning techniques (such as boosting, bagging, and RF) have got a great interest since they are robust to noise and more accurate than single classifiers. RF is a collection of tree structure classifiers.
For an RF that consists of N trees, the prediction of the class label c of a case x by majority voting is made by using the following equation:
where hn is the nth tree of the RF, and I is the indicator function.
7.5.3 Logistic regression (LR)
The LR [40] is a linear model used for classification problems. LR measures the relationship among the response (dependent) variable and one or more explanatory (independent) variables for a given dataset that indicates the significance and strength of the impact of the explanatory variables on the response variable. Typically, LR estimates probabilities using the logistic function, also known as the sigmoid function, which is given by [41]:
where e is the natural logarithm base, L is the curve’s maximum value, y0 is the y-value of the sigmoid’s midpoint, and k is the logistic growth rate or steepness of the curve.
7.5.4 K-nearest neighbor (KNN)
The KNN classifier is a nonparametric instance-based classifier [42]. This algorithm is based on the nearest neighborhood estimation. The new cases are classified on the basis of similarity measure which is the distance metric. In KNN, the K represents the number of nearest neighbor data values. Then, a similar instance is determined using the Euclidean distance formula [43].
7.6 Performance evaluation
The performance of the proposed method can be measured using well-known evaluation metrics—the accuracy of the classification, precision, recall, and F1 scores. These metrics are based on a “confusion matrix” that includes true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) [44, 45].
8 Experimental results and analysis
In this section, we have conducted experiments to assess the performance of the ML framework (MHF_HCV) for HCV disease diagnosis among HCWs in Egypt. As mentioned, the real-world data for HCV were used for MHF_HCV framework construction. We are conducting our experiments on a 3 GHz i5 computer with a 4 GB main memory and 64-bit Windows 7 operating system. The experiment is carried out using the python programming language.
Initially, the focus of the first part of this section is on measuring the effect of using different data splitting techniques on the performance of classification techniques, while in the second part, we focus on applying the wrapper feature selection method and evaluating the quality of the selected features by conducting a set of experiments. Finally, in the third part, we quantify the impact of parameter tuning on learning techniques.
8.1 Testing different data splitting methods
The performance ML model depends significantly on the data quality and the data strategy [46]. Therefore, assessment of the influence of data splitting on ML models’ performance has a high significance, which will pave the way to select a suitable data splitting method for better ML-based modeling. We conducted a comparative analysis of different data partitioning methods on real-world data for HCV using all features. Data splitting methods used in this study are k-fold cross-validation and random splitting using train–test splits method. Using the random splitting method, the dataset was divided into two parts, with different ratios: 70: 30 and 60:40 train/test split, as shown in Fig. 2a, in contrast performing k-fold cross-validation using k = 5 and k = 10, as shown in Fig. 2b.
Table 3 and Fig. 3 show the performance results for all classifiers using two data partitioning methods.
When applying classification techniques using the k-fold cross-validation (k = 10), their performance is better in most cases as shown in Table 3. We can also observe that the best performances occur using tenfold cross-validation with the RF classifier. Therefore, in this study, the tenfold cross-validation method for splitting the HCV dataset was found as the best option for ML modeling as shown in Fig. 3.
8.2 Influence of feature selection on the performance of the ML models
To examine the effect of feature selection on the classifier’s efficiency for the HCV dataset, we conducted a well-known classification technique: NB, RF, KNN, and LR. Classification techniques are used for HCV prediction in two scenarios as shown in Fig. 4: (1) without applying the proposed feature selection method and (2) with applying the SFS feature selection method. The evaluation metrics are listed in Sect. 4.6 used to measure the performance of classification techniques. The default parameters for each classification technique were used. The experiments were done by using a tenfold cross-validation method to select the best feature subset.
As mentioned before, the wrapper feature selection approach depends on the classification model. The feature selection approach, SFS, wrapped with different classifiers, namely NB, RF, KNN, and LR. Table 4 shows the number of features was selected when SFS covered with other classifiers.
As shown in Table 4, although the number of features is fixed for each classifier, the selected features are different. This is because the SFS feature selection method uses the classifier as an induction algorithm to select the features that achieve the best performance for each classifier. After that, the selected features are used for classification of testing samples. As shown in Table 5, the performance of various classification techniques is compared when using the first and second scenarios in terms of F1-scores, classification accuracy, recall, and precision. The highest values achieved are highlighted in bold format. The default parameters for each classification technique were used. The experiments were done using a tenfold cross-validation to evaluate the classifier’s performance.
From Table 5, when applying the classification technique using the first scenario with all features in the HCV dataset, we noticed that RF achieves higher performance with 94.06% accuracy, 63.71% F1-scores, 67.62% precision, and 61.95% recall.
In the second scenario, this experiment evaluates the effectiveness of applying classification techniques with only features selected using the SFS feature selection method shown in Table 4 and compares it with the first scenario.
In Table 5, we noticed that applying the classification using the second scenario can improve the performance of classifiers in terms of F1 scores, precision, recall, and accuracy, indicating that the features selected by the SFS method are more effective.
Although applying the RF algorithm with and without the SFS feature selection method has the same accuracy, the RF algorithm using the second scenario has achieved the same accuracy with a smaller set of features compared to the original set of features. Moreover, it has higher F1-scores (71.33%) and recall (84.52%) values.
These results also showed that the RF classifier using the second scenario has the best performance using only five features out of all features of the HCV dataset.
Figure 5 indicates the model’s learning time in seconds when using or not using the SFS feature selection technique for the HCV dataset.
From Fig. 5, we observed that classifiers using the second scenario consumed less time to build the model than the first scenario. Further, the results show that the NB algorithm consumes less time to create a model, while RF consumes a longer time.
The RF classifier using a reduced dataset had the best performance for the HCV dataset. While it took more time to build the model, it still less than the time required to build a model using the original dataset.
8.3 Influence of the parameter tuning on the learning algorithm
Parameter tuning is necessary because default values may not be appropriate and do not give the best results for all individual tasks [47]. As we inferred, the RF classifier, applying SFS feature selection, was the best-performed classifier. To optimize the classifier performance, we tuned its hyperparameters. As shown in Fig. 6, we used the grid search technique which tests a set of hyperparameters to determine the best parameter values for a given task based on validation accuracy. This is a computationally complex process than simply using the default parameter values for the model.
In this study, we used the following hyperparameters for the RF classifier:
-
n_estimators: number of trees in the forest.
-
max_depth: maximum depth of each tree.
-
max_features: The max_features parameter specifies the size of the random subsets of features to consider when splitting a node.
-
Criterion: The function to measure the quality of a split.
Table 6 shows the best hyperparameter values for the RF classifier. Alternatively, we utilized default hyperparameter values for other classifiers. In NB classifier, we used the default hyperparameter values such that the type of features, the number of features, the number of classes, and the size of the dataset also considered while building a Naive Bayes classifier in Table 4. In LR models, the hyperparameters performance, commonly used techniques for this, are grid search with tenfold cross-validation. The penalty used in LR = 12, c = 1, and verbose = 0. In KNN, we utilized k = 5, weights = “uniform”, algorithm = “auto”.
Once the best parameters were found, we tested the effectiveness of using hyperparameter tuning on the classification performance: first, we retrain the RF classifier with the full dataset using the new hyperparameter values. Then, we apply the SFS feature selection method wrapped with the RF classifier using the new parameter values to select the best feature subset. To evaluate the performance of RF classifier, the experiments were performed using a 10 cross-validation. Furthermore, new hyperparameter values are determined, as shown in Table 6.
Table 7 shows the effect of using hyperparameter tuning on the classification performance.
From Table 7, it is clear that the hyperparameter tuning improves the RF classifier performance. When using the SFS feature selection method wrapped with the RF classifier using the new parameters, the classification accuracy is improved using only five features out of all features of the HCV dataset.
9 Discussion and benefits of the research
In this paper, we proposed an ML framework, MHF_HCV, for HCV disease diagnosis among HCWs in Egypt. This study aims to work on methods to enhance the accuracy of ML classification methods to diagnose hepatitis C using real-world data for HCV. To achieve this, first, we measured the influence of the data splitting technique on classifiers’ performance. Then, we used an SFS-based wrapper feature selection approach to find an optimal feature subset that influences the class discovery process and improves the classification accuracy. After that, we test the performance of the classifiers using all attributes and selected features separately to compare the achieved accuracy and evaluate the quality of the selected features in improving the classification accuracy. Finally, we quantify the impact of parameter tuning on learning techniques. The results of the experiments presented in this work, which answer the Research questions, can be summarized as follows:
-
This paper proposes a prediction framework based on ML approaches to predict Hepatitis C Virus (HCV) among healthcare workers (HCWs) in Egypt. Thus, we applied a real-world data from the National Liver Institute (NLI), founded at Menoufiya University (Menoufiya, Egypt) which investigate the effectiveness of the proposed model.
-
When applying classification techniques using different splitting methods, we found that the tenfold cross-validation method for splitting the HCV dataset achieved best option for ML modeling and the overfitting problem is tackled.
-
When comparing classification techniques with and without the SFS feature selection method, we found that the features selected by SFS improved the performance of the classification techniques. Also, the RF classifier with the SFS feature selection method has the best performance using only five features out of all features of the HCV dataset.
-
RF classifier using a reduced dataset had the best performance for the HCV dataset; while it took more time to build the model, it is still less than the time required to build a model using the original dataset.
-
When adjusting the hyperparameter of the RF classifier, the performance of the RF classifier was improved. Also, when the SFS feature selection method wrapped with the RF classifier using new parameters, the classification accuracy was improved using only 4 features.
-
SFS feature selection method is a good choice for a feature selection method because of its simplicity, efficiency, and effectiveness. It can handle high-dimensional data well and it can handle imbalanced data well. It is also easy to interpret and understand which makes it a good choice for many machine learning tasks.
As concluding observations, the RF classifier using tenfold cross-validation yielded the highest effectiveness with parameter tuning and SFS feature selection, achieving an accuracy score of 94.88. Also, the work presented in this research has proved the objectives of this study where the results obtained demonstrate its effectiveness.
10 Threads and limitations
Although the proposed framework achieved superior performance in diagnosing HCV disease, it still has some limitations. The first one is that the sample selected for this study was specifically for Egyptian HCWs, working in a high-risk environment in NLI and not for HCV patients in general. The second limitation is the size of the HCV data set, which consists of 859 patients. Using a large HCV dataset to train the model could improve the performance of the proposed HCV framework. The current study used only 11 features that represent the results of laboratory tests required to diagnose HCV. Therefore, more features are required to give more details that may be useful in diagnosing newly infected cases of HCV. The motivations for selecting classifiers (NB, RF, KNN, LR) are presented as follows: Firstly, the mentioned methodologies achieved satisfactory results in terms of accuracy before and after feature selection. Secondly, due to the limited number of records of the applied dataset, the size consists of only 859 (patients) records. Third, we performed an experimental result based on different ML methods, and the results obtained could have been better compared with the selected NB, RF, KNN, and LR results. Machine learning techniques are more suited to small datasets than deep learning approaches, which need vast data to produce more accurate findings. We investigate the use of deep learning methods on huge datasets in future work.
11 Conclusion and future work
This paper proposes an ML framework based on NB, RF, KNN, and LR to classify and predict the infected patients with HCV for the enrolled features. SFS feature selection is presented to select the most significant features of the applied dataset. To manipulate the enrolled data without feature selection, we also tested the dataset by using all the features directly to the NB, RF, KNN, and LR without feature selection. Finally, we tested the impact of parameter tuning on learning techniques. The results indicate that there is a great enhancement of the accuracy obtained after feature selection using SFS with the new parameters in terms of accuracy. Moreover, the elapsed learning time is very law which means the speed of the learning process of the proposed framework. In future work, we plan to recommend a treatment protocol based on a genetic algorithm or DNA sequence analysis to help the specialist treat and handle the spread of HCV in Egypt.
References
WHO (2021) Hepatitis C. Httpswwwwhointnews-Roomfact-Sheetsdetailhepatitis-C
Mohamed AA, Elbedewy TA, El-Serafy M et al (2015) Hepatitis C virus: a global view. World J Hepatol 7:2676
Huang R, Rao H, Yang M et al (2020) Noninvasive measurements predict liver fibrosis well in hepatitis C virus patients after direct-acting antiviral therapy. Dig Dis Sci 65:1491–1500
Westermann C, Peters C, Lisiak B et al (2015) The prevalence of hepatitis C among healthcare workers: a systematic review and meta-analysis. Occup Environ Med 72:880–888
John GH, Kohavi R, Pfleger K (1994) Irrelevant features and the subset selection problem. Machine learning proceedings. Elsevier, Amsterdam, pp 121–129
Triantaphyllou E, Felici G (2006) Data mining and knowledge discovery approaches based on rule induction techniques. Springer, New York
Taz NH, Islam A, Mahmud I (2021) A comparative analysis of ensemble based machine learning techniques for diabetes identification. In: 2021 2nd International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST). IEEE, pp 1–6
Doğru A, Buyrukoğlu S, Arı M (2023) A hybrid super ensemble learning model for the early-stage prediction of diabetes risk. Med Biol Eng Comput 1–13
Hoofnagle JH, Di Bisceglie AM (1997) The treatment of chronic viral hepatitis. N Engl J Med 336:347–356
Jaeckel E, Cornberg M, Wedemeyer H et al (2001) Treatment of acute hepatitis C with interferon alfa-2b. N Engl J Med 345:1452–1457
Frank C, Mohamed MK, Strickland GT et al (2000) The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet 355:887–891
Antaki N, Craxi A, Kamal S et al (2010) The neglected hepatitis C virus genotypes 4, 5 and 6: an international consensus report. Liver Int 30:342–355
Burra P, Senzolo M, Adam R et al (2010) Liver transplantation for alcoholic liver disease in Europe: a study from the ELTR (European Liver Transplant Registry). Am J Transpl 10:138–148
Bruno S, Crosignani A, Facciotto C et al (2010) Sustained virologic response prevents the development of esophageal varices in compensated, Child-Pugh class A hepatitis C virus–induced cirrhosis. a 12-year prospective follow-up study. Hepatology 51:2069–2076
Bini EJ, Perumalswami PV (2010) Hepatitis B virus infection among American patients with chronic hepatitis C virus infection: prevalence, racial/ethnic differences, and viral interactions. Hepatology 51:759–766
Ali MMR, Helmy Y, Khedr AE, Abdo A (2018) Intelligent decision framework to explore and control infection of hepatitis C virus. International conference on advanced machine learning technologies and applications. Springer, New York, pp 264–274
Abd El-Salam SM, Ezz MM, Hashem S et al (2019) Performance of machine learning approaches on prediction of esophageal varices for Egyptian chronic hepatitis C patients. Inform Med Unlocked 17:100267
Nandipati SC, XinYing C, Wah KK (2020) Hepatitis C virus (HCV) prediction by machine learning techniques. Appl Model Simul 4:89–100
UCI-ML repository HCV UCI-ML repository. https://archive.ics.uci.edu/ml/datasets/Hepatitis+C+Virus+%28HCV%29+for+Egyptian+patients
Eliyahu S, Sharabi O, Elmedvi S et al (2018) Antibody repertoire analysis of hepatitis C virus infections identifies immune signatures associated with spontaneous clearance. Front Immunol 9:3004
Hashem S, ElHefnawi M, Habashy S et al (2020) Machine learning prediction models for diagnosing hepatocellular carcinoma with HCV-related chronic liver disease. Comput Methods Programs Biomed 196:105551
KayvanJoo AH, Ebrahimi M, Haqshenas G (2014) Prediction of hepatitis C virus interferon/ribavirin therapy outcome based on viral nucleotide attributes using machine learning algorithms. BMC Res Notes 7:1–11
Alade OM, Sowunmi OY, Misra S et al (2018) A neural network based expert system for the diagnosis of diabetes mellitus. In: Antipova T, Rocha Á (eds) Information technology science. Springer, Cham, pp 14–22
Abayomi-Alli OO, Damaševičius R, Maskeliūnas R, Abayomi-Alli A (2020) BiLSTM with data augmentation using interpolation methods to improve early detection of parkinson disease. In: 2020 15th conference on computer science and information systems (FedCSIS). pp 371–380
Ogundokun RO, Misra S, Sadiku PO et al (2022) Computational intelligence approaches for heart disease detection. In: Singh PK, Singh Y, Chhabra JK et al (eds) Recent innovations in computing. Springer, Singapore, pp 385–395
Salem H, Shams MY, Elzeki OM et al (2022) Fine-tuning fuzzy KNN classifier based on uncertainty membership for the medical diagnosis of diabetes. Appl Sci 12:950
Buyrukoğlu S (2021) New hybrid data mining model for prediction of Salmonella presence in agricultural waters based on ensemble feature selection and machine learning algorithms. J Food Saf 41:e12903
Buyrukoğlu G, Buyrukoğlu S, Topalcengiz Z (2021) Comparing regression models with count data to artificial neural network and ensemble models for prediction of generic Escherichia coli population in agricultural ponds based on weather station measurements. Microb Risk Anal 19:100171
Buyrukoğlu S (2021) Promising cryptocurrency analysis using deep learning. In: 2021 5th International symposium on multidisciplinary studies and innovative technologies (ISMSIT). IEEE, pp 372–376
Buyrukoğlu S, Savaş S (2022) Stacked-based ensemble machine learning model for positioning footballer. Arab J Sci Eng 1–13
Akbas A, Buyrukoglu S (2022) Stacking ensemble learning-based wireless sensor network deployment parameter estimation. Arab J Sci Eng 1–10
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Ijcai. Montreal, Canada, pp 1137–1145
Liu H, Cocea M (2017) Semi-random partitioning of data into training and test sets in granular computing context. Granul Comput 2:357–386
Chandrashekar G, Sahin F (2014) A survey on feature selection methods. Comput Electr Eng 40:16–28
Kohavi R, John GH (1997) Wrappers for feature subset selection. Artif Intell 97:273–324
Gutierrez-Osuna R (2002) Pattern analysis for machine olfaction: a review. IEEE Sens J 2:189–202
Langley P (1994) Selection of relevant features in machine learning. In: Proceedings of the AAAI fall symposium on relevance. pp 245–271
Gopal M (2019) Applied machine learning. McGraw-Hill Education, New York
Breima L (2010) Random forests. Mach Learn 45:5–32
Le Cessie S, Van Houwelingen JC (1992) Ridge estimators in logistic regression. J R Stat Soc Ser C Appl Stat 41:191–201
Shu J, Tang Y, Cui J et al (2018) Clear cell renal cell carcinoma: CT-based radiomics features for the prediction of Fuhrman grade. Eur J Radiol 109:8–12
Kumari R, Jose J (2011) Seizure detection in EEG using Biorthogonal wavelet and fuzzy KNN classifier. Elixir Hum Physiol 41:5766–5770
Altay O, Ulas M (2018) Prediction of the autism spectrum disorder diagnosis with linear discriminant analysis classifier and K-nearest neighbor in children. In: 2018 6th International symposium on digital forensic and security (ISDFS). IEEE, pp 1–4
Sokolova M, Japkowicz N, Szpakowicz S (2006) Beyond accuracy, F-score and ROC: a family of discriminant measures for performance evaluation. Australasian joint conference on artificial intelligence. Springer, New York, pp 1015–1021
Raschka S (2014) An overview of general performance metrics of binary classifier systems. ArXiv Preprint: http://arxiv.org/abs/14105330
Asteris PG, Apostolopoulou M, Armaghani DJ et al (2020) On the metaheuristic models for the prediction of cement-metakaolin mortars compressive strength. Metaheuristic Comput Appl 1(1):063
Hartmann J, Huppertz J, Schamp C, Heitmann M (2019) Comparing automated text classification methods. Int J Res Mark 36:20–38
Acknowledgements
Authors sincerely acknowledge Computer Science Department in Faculty of Science, Minia University, for the facilities and support.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
Heba and Tarek wrote the main manuscript text and Mahmoud prepared figures. All authors reviewed the manuscript and implement the prposed work
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mamdouh Farghaly, H., Shams, M.Y. & Abd El-Hafeez, T. Hepatitis C Virus prediction based on machine learning framework: a real-world case study in Egypt. Knowl Inf Syst 65, 2595–2617 (2023). https://doi.org/10.1007/s10115-023-01851-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-023-01851-4