Abstract
Following the observation of an increased number of isolation of OXA-23- and ArmA-producing Acinetobacter baumannii at the national level, our aim was to evaluate whether some given clone(s) might actually be spreading and/or emerging in Switzerland. To evaluate this possibility, our study investigated and characterized all A. baumannii isolates harboring both the blaOXA-23 and armA genes that had been collected at the Swiss National Reference Center for Emerging Antibiotic Resistance (NARA) from 2020 to 2021. Most isolates were obtained from infections rather than colonization with the majority being obtained from respiratory specimens. Pulsed-field gel electrophoresis (PFGE) analysis of 56 isolates identified nine profiles. Then, whole-genome sequencing that was performed on a subset of 11 isolates including at least one representative isolate of each PFGE profile identified three STs; one each of ST25 and ST1902, and nine ST2 (a member of Global Clone 2 (GC-2). The blaOXA-23 gene was always found embedded within Tn2006 structures, as commonly described with GC-2 (ST2) isolates. Susceptibility testing showed that most of those isolates, despite being highly resistant to all carbapenems and all aminoglycosides, remained susceptible to colistin (94.6%), sulbactam-durlobactam (87.5%), and cefiderocol (83.9% or 91.1% according to EUCAST or CLSI breakpoints, respectively). Overall, this study identified that the A. baumannii co-producing OXA-23 and ArmA are increasing in incidence in Switzerland, largely due to the dissemination of the high-risk GC-2. This highlights the importance of the monitoring of such MDR A. baumannii strains, in order to contribute to reduce their potential further spread.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carbapenem-resistant Acinetobacter baumannii (CRAB) present an urgent global public health threat, due to both the limited treatment options for infections caused by CRAB, which are usually multidrug resistant, and the increasing reports of infections [1]. Several groups of carbapenem-hydrolysing class D ß-lactamases have been described in Acinetobacter spp., but OXA-23 is the most frequently reported acquired class D carbapenemase [1]. OXA-23 was initially described in 1995 [2], from an A. baumannii isolate, collected in Scotland in 1985, and the sequence was later published in 2000 [3]. In this study, the blaOXA-23 gene was found to be encoded on a transferable plasmid, with the corresponding enzyme exhibiting significant carbapenemase activity [3]. Following its identification, OXA-23-producing A. baumannii have since been extensively reported worldwide, often as a cause of nosocomial outbreaks, and mainly associated with the globally disseminated high-risk clones belonging to Global Clone 1 (GC-1) and Global Clone 2 (GC-2) (also referred to as International Clones) [1, 4, 5]. Those OXA-23 producers are most often resistant to many other antibiotic families, and in particular to fluoroquinolones, tetracycline, and chloramphenicol. They are also commonly resistant to different aminoglycosides, due to the acquisition of aminoglycoside-modifying enzymes. Furthermore, many of them exhibit pan-resistance to all aminoglycosides, including plazomicin [6], resulting from the acquisition of 16S rRNA methylases, among which ArmA is the most prevalent enzyme.
The current treatment options for CRAB infections include aminoglycosides, polymyxins, tigecycline, minocycline, cefiderocol, and might soon include a novel ß-lactam/ß-lactamase inhibitor combination, namely sulbactam-durlobactam [7, 8]. Co-resistance to carbapenems and all aminoglycosides in Acinetobacter spp. has been reported in numerous studies and has been predominantly associated with the co-carriage of blaOXA-23 and armA genes among isolates belonging to GC-2 [9, 10].
Having observed an increased number of isolation of OXA-23- and ArmA-producing A. baumannii at the national level, our aim was to evaluate whether some given clone(s) might actually be spreading and/or emerging in Switzerland. To evaluate this possibility, our study investigated and characterized all A. baumannii isolates harboring both the blaOXA-23 and armA genes that had been collected at the Swiss National Reference Center for Emerging Antibiotic Resistance (NARA) from 2020 to 2021.
Materials and methods
Bacterial isolates, identification, susceptibility testing and typing
Seventy-five isolates were submitted to the NARA reference laboratory from hospitals and clinics throughout Switzerland, over a 2-year period, from Jan 2020 to Dec 2021. Species identification was determined using MALDI-TOF MS (Bruker Microflex LT, Bruker Daltonik GmbH, Bremen, Germany) and UriSelect 4 agar (Bio-Rad, https://www.bio-rad.com). Susceptibility testing was performed by either disk diffusion or broth microdilution, and results were interpreted in accordance with EUCAST guidelines, as well as CLSI guidelines in the case of cefiderocol [11, 12]. All isolates were subject to the Rapidec Carba NP test (bioMérieux, La Balme-les-Grottes, France) and then to NG-Test CARBA 5 test (NG Biotech), according to the manufacturer’s instructions. The occurrence of the blaOXA-23 and armA genes was confirmed by PCR and subsequent Sanger sequencing. Isolates were typed by PFGE using ApaI-digested genomic DNA as previously described [13].
Whole-genome sequencing (WGS) and analyses
WGS was performed on a subset of eleven isolates, selected to be representative of different PFGE profiles. All eleven isolates were sequenced using the Illumina (short read) platform, and six isolates were selected for Nanopore Technologies (ONT) (long read) sequencing, as previously described [14]. Illumina reads were trimmed using Trimmomatic [15], and ONT reads were corrected and trimmed using Canu [16], and assembled using Flye [17]. Flye assemblies were then polished with Illumina reads using CLC Genomics Workbench (Qiagen). Sequence types and the presence of resistance genes were identified using MLST version 2.0, and ResFinder version 4.1 on the Center for Genomic Epidemiology platform (https://cge. cbs.dtu.dk); contigs were annotated using Prokka [18].
Sequence data from this study was submitted to the National Center for Biotechnology Information’s Sequence Read Archive (BioProject no. PRJNA891672).
Results and discussion
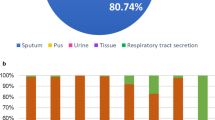
Between 2017 and 2021, a total of 188 carbapenem-resistant A. baumannii were submitted to the NARA and around two-thirds (131/188; 69.7%) produced an OXA-23 carbapenemase. The remaining 57 CRAB harbored diverse carbapenem-resistance mechanisms including the production of other carbapenemases, predominantly NDM-type and OXA-40 enzymes. Five isolates also harbored a second carbapenemase gene, three with blaNDM-1 and two with blaOXA-40. Within these 131 OXA-23 producers, most (n = 98, 74.8%) also harbored the armA gene. The majority of these OXA-23 and ArmA co-producing isolates (75/98; 76.5%) were submitted over a 2-year period, from Jan 2020 to Dec 2021. Notably, the incidence of both carbapenem-resistant A. baumannii and OXA-23-producing A. baumannii submitted to NARA have increased significantly, year on year, over this time period (Fig. 1). Isolates in this study were mainly obtained from infections rather than colonizations (Table 1) in over half of all cases (CRAB; 57.5%, OXA-23 producers; 56.7%, OXA-23- + ArmA-producers; 62.2%), with the majority obtained from respiratory specimens.
PFGE typing of 56 non-duplicate (by patient and specimen type) isolates from within the studied collection, selected to be geographically representative and submitted to NARA between Jan 2020 and Dec 2021, identified nine distinct PFGE profiles (designated A to I). The 56 isolates were predominantly obtained from either feces (16/56; 28.6%) or respiratory specimen types (13/56; 23.2%). MLST analysis, according to the Pasteur scheme [5], was performed on eleven isolates from six Swiss cantons, with at least one representative of each PFGE profile. Six of these, representing of 5 different PFGE profiles, were selected for further analysis by long-read WGS. The characteristics of these isolates are shown in Table 2. Among the eleven isolates, three STs were found comprising ST2 (n = 9) (corresponding to GC-2) [1, 19], and one each of ST25 and ST1902, respectively. Among the nine identified pulsotypes, isolates were distributed as follows (no./no. Swiss Cantons): A (n = 10/7), B (n = 15/6), C (n = 7/5), D (n = 7/5), E (n = 10/6), F (n = 4/3), and one representative each of pulsotypes G, H, and I. Analyses of the six isolates subjected to long-read WGS identified that the blaOXA-23 and armA genes were located on the chromosome in six and four isolates respectively. Interestingly, two copies of the blaOXA-23 gene were present in 5 out of those 6 isolates. Multiplication of blaOXA-23 in the A. baumannii chromosome has been reported previously [19, 20], although there are contrasting reports about whether or not this has any effect on carbapenem MICs [19, 20]. Of note, the occurrence of such blaOXA-23 multicopy is likely to be underestimated due to the relative inability of short-read sequencing to detect such duplications upon WGS. The blaOXA-23 genes were all found embedded within Tn2006 structures, as previously described with GC-2 (ST2) isolates [21].
Apart from being categorized to be resistant to all aminoglycosides and carbapenems (meropenem, imipenem), susceptibility testing revealed susceptibility levels to colistin (COL) at 94.6%, to minocycline at 23.2%, to sulbactam at 1.8%, to sulbactam-durlobactam (SUL-DUR) at 87.5%, and to cefiderocol (FDC) at 83.9% or 91.1% (according to EUCAST [11] or CLSI [12] breakpoints, respectively) (Table 3). This underscores that such isolates may be considered as susceptible to very few therapeutic options, namely COL, SUL-DUR, and FDC, respectively.
Overall, this study identified that the A. baumannii co-producing OXA-23 and ArmA are increasing in incidence in Switzerland, and this is largely due to the dissemination of the high-risk GC-2. In particular, ST2 strains were mainly found all over the country, suggesting that this latter clonal background might be considered as a “successful” one. Interestingly, widespread dissemination of ST2 strains co-producing OXA-23 and ArmA has already been evidenced in other countries, such as Bangladesh [22], Yemen [23], Vietnam [24], and Latvia [25].
Prevention of dissemination of A. baumannii strains in general, and of those multidrug-resistant strains particular, relies mainly on the quality of infection prevention and control interventions [26]. Noteworthy, the occurrence of such multidrug-resistant A. baumannii isolates remained quite low in Switzerland during a long period, with only 58 carbapenem-resistant isolates being reported all over the country from 2005 to 2016 from institutions representing 70% of all hospitalized patients and one-third of all ambulatory practitioners in the country [27]. The issue of early detection of such isolates when colonizing hospitalized patients is surely a major concern. Among the recently developed rapid diagnostic tests for the identification of carbapenem resistance in A. baumannii, the use of the Rapid ResaImipenem/Acinetobacter NP test is a fast and reliable approach, since it allows to categorize the status of either susceptibility or resistance to carbapenems of any A. baumannii isolates within 2 h 30 min, with 100% sensitivity and specificity [28].
Through our study, a high rate of ArmA-producing strains has been evidenced. Also noteworthy is therefore the possibility to screen for 16S rRNA methylase producers directly from rectal swabs in order to eventually reinforce the infection control measures. This can now be considered in clinical laboratories by using the so far unique screening medium for that purpose, namely the SuperAminoglycoside medium [29]. Overall, it is therefore important to highlight that strict monitoring of such MDR A. baumannii strains is needed, in order to contribute to reduce their potential further spread.
References
Hamidian M, Nigro SJ (2019) Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom 5(10):e000306. https://doi.org/10.1099/mgen.0.000306
Scaife W, Young H-K, Paton RH et al (1995) Transferable imipenem-resistance in Acinetobacter species from a clinical source. J Antimicrob Chemother 36:585–587
Donald HM, Scaife W, Amyes SGB et al (2000) Sequence analysis of ARI-1, a novel OXA ß-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother 44(1):196–199. https://doi.org/10.1128/AAC.44.1.196-199.2000
Zarrilli R, Pournaras S, Giannouli M et al (2013) Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41(1):11–19. https://doi.org/10.1016/j.ijantimicag.2012.09.008
Diancourt L, Passet V, Nemec A et al (2010) The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5(4):e10034. https://doi.org/10.1371/journal.pone.0010034
Taylor E, Sriskandan S, Woodford N et al (2018) High prevalence of 16S rRNA methyltransferases among carbapenemase-producing Enterobacteriaceae in the UK and Ireland. Int J Antimicrob Agents 52:278–282. https://doi.org/10.1016/j.ijantimicag.2018.03.016
Abdul-Mutakabbir JC, Griffith NC, Shields RK et al (2021) Contemporary perspective on the treatment of Acinetobacter baumannii infections: insights from the Society of Infectious Diseases Pharmacists. Infect Dis Ther 10(4):2177–2202. https://doi.org/10.1007/s40121-021-00541-4
Karlowsky JA, Hackel MA, McLeod SM et al (2022) In vitro activity of sulbactam-durlobactam against global isolates of Acinetobacter baumannii-calcoaceticus Complex collected from 2016 to 2021. Antimicrob Agents Chemother 20;66(9):e0078122. https://doi.org/10.1128/aac.00781-22.
Camargo CH, Yamada AY, Nagamori FO et al (2022) Clonal spread of ArmA- and OXA-23-coproducing Acinetobacter baumannii International Clone 2 in Brazil during the first wave of the COVID-19 pandemic. J Med Microbiol 71(4). https://doi.org/10.1099/jmm.0.001509
Nafplioti K, Galani I, Angelidis E et al (2020) Dissemination of international clone II Acinetobacter baumannii strains coproducing OXA-23 carbapenemase and 16S rRNA methylase ArmA in Athens. Greece. Microb Drug Resist 26(1):9–13. https://doi.org/10.1089/mdr.2019.0075
European committee on antimicrobial susceptibility testing. Clinical breakpoint table v. 12.0. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf. Accessed 21 Dec 2022
CLSI (2022) Performance standards for antimicrobial susceptibility testing, M100, 32nd edn. Clinical and Laboratory Standards Institute, Wayne, PA
Seifert H, Dolzani L, Bressan R et al (2005) Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43(9):4328–4335. https://doi.org/10.1128/JCM.43.9.4328-4335.2005
Findlay J, Perreten V, Poirel L et al (2022) Molecular analysis of OXA-48-producing Escherichia coli in Switzerland from 2019 to 2020. Eur J Clin Microbiol Infect Dis 41(11):1355–1360. https://doi.org/10.1007/s10096-022-04493-6
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Koren S, Walenz BP, Berlin K et al (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27(5):722–736
Evans BA, Amyes SG (2014) OXA β-lactamases. Clin Microbiol Rev 27(2):241–263. https://doi.org/10.1128/CMR.00117-13
Seeman T (2014) Prokka: rapid prokaryotic genome annotation. Bioinf 30:2068–2069
Hua X, Shu J, Ruan Z et al (2016) Multiplication of blaOXA-23 is common in clinical Acinetobacter baumannii, but does not enhance carbapenem resistance. J Antimicrob Chemother 12:3381–3385
Zhang Q, Hua X, Ruan Z et al (2018) Revisiting the contribution of gene duplication of blaOXA-23 in carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother 1:250–252. https://doi.org/10.1093/jac/dkx339
Mugnier PD, Poirel L, Naas T et al (2010) Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis 16(1):35–40. https://doi.org/10.3201/eid1601.090852
Farzana R, Swedberg G, Giske CG et al (2022) Molecular and genetic characterization of emerging carbapenemase-producing Acinetobacter baumannii strains from patients and hospital environments in Bangladesh. Infect Prev Pract 27 4(2):100215
Bakour S, Alsharapy SA, Touati A et al (2014) Characterization of Acinetobacter baumannii clinical isolates carrying blaOXA-23 carbapenemase and 16S rRNA methylase armA genes in Yemen. Microb Drug Resist 20(6):604–609
Tada T, Miyoshi-Akiyama T, Shimada K et al (2015) Dissemination of clonal complex 2 Acinetobacter baumannii strains co-producing carbapenemases and 16S rRNA methylase ArmA in Vietnam. BMC Infect Dis 15(15):433
Saule M, Samuelsen Ø, Dumpis U et al (2013) Dissemination of a carbapenem-resistant Acinetobacter baumannii strain belonging to international clone II/sequence type 2 and harboring a novel AbaR4-like resistance island in Latvia. Antimicrob Agents Chemother 57(2):1069–1072
Tomczyk S, Zanichelli V, Grayson ML et al (2019) Control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in healthcare facilities: a systematic review and reanalysis of quasi-experimental studies. Clin Infect Dis 68(5):873–884
Ramette A, Kronenberg A, the Swiss Centre for Antibiotic Resistance (ANRESIS) (2018) Prevalence of carbapenem-resistant Acinetobacter baumannii from 2005 to 2016 in Switzerland. BMC Infect Dis 18(1):159
Nordmann P, Sadek M, Tinguely C et al (2021) Rapid ResaImipenem/Acinetobacter NP test for detection of carbapenem susceptibility/resistance in Acinetobacter baumannii. J Clin Microbiol 59(6):e03025–e03020
Nordmann P, Mazé A, Culebras E et al (2018) A culture medium for screening 16S rRNA methylase-producing pan-aminoglycoside resistant Gram-negative bacteria. Diagn Microbiol Infect Dis 91(2):118–122
Acknowledgements
We thank the following colleagues for providing us with the A. baumannii clinical strains: M. Oberle, C. Ottiger (Aargau); O. Dubuis, S. Graf (Basel-Landschaft); A. Egli, D. Goldenberger (Basel Stadt); K. Burren, C. Casanova, S. Droz, S. Thiermann, R. Troesch M. Verena, (Bern); D. Bandera, V. Deggim, S. Pfister, S. Trachsel (Fribourg); N. Liassine, G. Renzi (Geneva); C. Guler (Grisons); L. Monnerat (Jura); I. Dietrich, P. Friderich, I. Mitrovic, S. Pranghofer, B. Suterbuser (Lucerne); K. Vidakovic (Schaffhausen); M. Ritzler, S. Sieffert (St Gallen); K. Herzog (Thurgau); V. Gaia (Ticino); S. Emonet, M. Eyer, L. Tissieres (Valais); C. Andreutti, M. Corthesy, M. Dessauges, M. Maitrejean, M. Rosselin, C. Vogne (Vaud); V. Bruderer, G. Eich, E. Gruner, F. Kaeppeli, M. Kuegler, K. Lucke, P. Minkova, S. Mancini (Zurich).
Funding
Open access funding provided by University of Fribourg This work was financed by the University of Fribourg, Switzerland, the Swiss National Science Foundation (project FNS-407240_177381), and by the NARA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Findlay, J., Nordmann, P., Bouvier, M. et al. Dissemination of ArmA- and OXA-23-co-producing Acinetobacter baumannii Global Clone 2 in Switzerland, 2020–2021. Eur J Clin Microbiol Infect Dis (2023). https://doi.org/10.1007/s10096-023-04643-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10096-023-04643-4