Abstract
This study determined the carriage rates and antimicrobial resistance (AMR) genes of enterococci from nasotracheal samples of three healthy animal species and in-contact humans. Nasal samples were collected from 27 dog-owning households (34 dogs, 41 humans) and 4 pig-farms (40 pigs, 10 pig-farmers), and they were processed for enterococci recovery (MALDI-TOF–MS identification). Also, a collection of 144 enterococci previously recovered of tracheal/nasal samples from 87 white stork nestlings were characterized. The AMR phenotypes were determined in all enterococci and AMR genes were studied by PCR/sequencing. MultiLocus-Sequence-Typing was performed for selected isolates. About 72.5% and 60% of the pigs and pig-farmers, and 29.4% and 4.9%, of healthy dogs and owners were enterococci nasal carriers, respectively. In storks, 43.5% of tracheal and 69.2% of nasal samples had enterococci carriages. Enterococci carrying multidrug-resistance phenotype was identified in 72.5%/40.0%/50.0%/23.5%/1.1% of pigs/pig-farmers/dogs/dogs’ owners/storks, respectively. Of special relevance was the detection of linezolid-resistant enterococci (LRE) in (a) 33.3% of pigs (E. faecalis-carrying optrA and/or cfrD of ST59, ST330 or ST474 lineages; E. casseliflavus-carrying optrA and cfrD); (b) 10% of pig farmers (E. faecalis-ST330-carrying optrA); (c) 2.9% of dogs (E. faecalis-ST585-carrying optrA); and (d) 1.7% of storks (E. faecium-ST1736-carrying poxtA). The fexA gene was found in all optrA-positive E. faecalis and E. casseliflavus isolates, while fexB was detected in the poxtA-positive E. faecium isolate. The enterococci diversity and AMR rates from the four hosts reflect differences in antimicrobial selection pressure. The detection of LRE carrying acquired and transferable genes in all the hosts emphasizes the need to monitor LRE using a One-Health approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enterococcus spp. are commensals and predominantly found in the intestinal habitat, but they might be translocated to other animal tissues or organs [1]. Among the over 50 different enterococci species, Enterococcus faecium (E. faecium) and Enterococcus faecalis (E. faecalis) constitute most of the gastrointestinal tract (GI) enterococci communities in humans [2]. However, in livestock, E. faecium, E. cecorum, E. faecalis and, to some extent, E. hirae predominate [3]. In contrast, E. mundtii and E. casseliflavus are commonly found in plant and environmental samples [2, 4]. Moreover, the ecologic-epidemiology of E. faecalis and E. faecium has shown animal food (such as pork) and the environment (sewage, soil, and water) as common colonized items [4, 5].
Enterococci are very hardy organisms; they can sustain various adverse conditions and survive for several months in the environment [2]. These attributes make enterococci challenging to control once established in a hospital, household, or pig-farm setting. Enterococci are also suitable as important key indicator bacteria for veterinary and human resistance surveillance systems [4, 6]. Enterococci can easily acquire antimicrobial resistance (AMR) through mutations or the acquisition of antimicrobial resistance genes (ARGs), included in plasmids and transposons [7].
The livestock industry plays an important role in the transmission of multidrug-resistant (MDR) enterococci isolates due to the close interaction between farmers, livestock, and the farm environment [7, 8]. However, a new European Union law now prohibits the use of antimicrobial agents as prophylactics in feeds [9]. The pervasive selection of resistant bacteria in livestock facilitates the persistence and dissemination of MDR isolates to other animals and humans [7]. The spread of such MDR isolates can occur through direct (consumption/handling of contaminated food, direct contact with farmers/veterinarians) or indirect routes (animal waste handling, contaminated groundwater or surfaces) [10].
Among antimicrobial resistance transmission, it is important to remark that the widespread use of chloramphenicol in the past for farm animals has been able to select bacteria resistant to this agent and maybe to other clinically important antibiotics (by co-selection) [11]. Linezolid is one of the most important treatment options for severe infections by enterococci, including vancomycin-resistant enterococci (VRE). Increasing reports on linezolid-resistant enterococci (LRE) detected throughout the agricultural sector including poultry, pigs and cattle indicate that linezolid resistance might be co-selected by the use of chloramphenicol in livestock, with potentially serious consequences for public health [12,13,14].
Interestingly, a study on farm animals found limited sharing of isolates and resistance genes between livestock and humans, except for some pig isolates that were genetically related to hospital-associated isolates [15]. By contrast, dogs may be a reservoir of hospital-associated E. faecium clones and may form a higher risk for zoonotic transfer to humans [16]. Occupational contact with livestock plays a major role in certain professions, such as veterinarians, slaughterhouse workers or farmers [17, 18]. However, in the case of pets, not only veterinarians but also animal owners are at risk of acquiring MDR zoonotic bacterial pathogens [19]. It has been observed as enterococci, in particular E. faecalis and E. faecium, have zoonotic potential [20]. Dogs are companion animals that have been in close contact with humans since ancient times, which increases the likelihood of the transmission of bacteria between these animals and their owners [21].
Finally, wildlife has been considered key players in the carriage and transmission of AMR as many of them, especially the migratory birds (such as storks), could be dynamic and move along distance across a variety of natural environments, landfills, and livestock farms [22]. Migratory birds occasionally come in contact with antibiotic residues in livestock carcasses or manure and they could carry and disseminate AMR bacteria such as Enterococcus of public health concerns [22].
Studies on LRE of animal origin are available in some European countries, including Spain. However, studies reported on nasal LRE carriage are very rare. Especially among Enterococcus species other than E. faecalis and E. faecium. This prospective comparative study sought to determine the prevalence of nasal carriage of Enterococcus sp. and the molecular characterization of isolates in a population of healthy dogs, dog-owning households, pigs and pig-farmers, and storks in Spain, with special focus on linezolid resistance characterization.
Material and methods
Study participants’ descriptions and samples analyses
Nasal samples were obtained during 2021–2022 from animals and humans of the following origins: (a) 27 dog-owning households (34 dogs, 41 humans) in La Rioja region (Spain); (b) 40 pigs of 4 farms (A–D) comprising 10 pigs from each farm from the Aragon region (Spain), and 10 workers of the pig-farms (2, 3, 2 and 3 humans in farms A–D, respectively). The nasal samples were obtained using sterile swabs with conservation media (Amies, City, Country), and were used for enterococci recovery as below indicated. Moreover, a collection of 144 enterococci previously recovered from nasal and tracheal samples of 87 nestling white storks [23] were included in this study for phenotypic and genotypic characterization of antimicrobial resistance and molecular typing; these nestlings corresponded to four different colonies of Ciudad Real region (Center-South of Spain) with parent storks foraging on different habitats (colonies 1 and 2: located in natural habitat; colonies 3 and 4: foraging in landfills). The storks isolates were of the following species (number of isolates): E. faecalis (78), E. faecium (44), E. cecorum (8), E. casseliflavus (5), E. gallinarum (2), E. durans (2), E. hirae (1) and E. canis (n = 1).
Collected nasal and tracheal samples were enriched in brain heart infusion broth (BHI; Condalab, Madrid, Spain) supplemented with 6.5% NaCl and incubated for 24 h at 37 °C. After overnight incubation, different dilutions of the broth culture were carefully dispensed onto blood agar (BioMerieux) and ChromAgar LIN (CHROMagar™ LIN, Paris, France) plates and incubated for 24 h at 37 °C, for enterococci recovery. The CHROMAgar™ medium has been previously shown to have high sensitivity and specificity on pure linezolid resistant enterococci and staphylococci isolates [24]. After overnight growth, 2 to 5 different colonies per sample with the morphology of enterococci were randomly selected and identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF; Bruker Daltonics, Bremen, Germany) using the standard extraction protocol recommended by Bruker.
All sampling procedures were performed following all applicable international, national, and/or institutional guidelines for human samples experiments (as described in the revised Helsinki declaration) and for ethical use of animals, specifically directive 2010/63/EU and Spanish laws 9/2003 and 32/2007, and RD 178/2004 and RD 1201/2005. All procedures were approved by the ethical committees of the University of La Rioja, the University of Zaragoza and the University of Castilla La Mancha of Spain.
Enterococci DNA extraction
The DNA extraction of enterococci isolates of all origins was performed using InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, USA), according to the manufacturer’s instructions. Briefly, pure and fresh isolated colonies were suspended in 1000 μL of sterile Milli-Q water, thoroughly mixed by vortex, and centrifuged at 12,000 revolutions per minute for 3 min. The supernatant was carefully eliminated and 20 μL of InstaGene matrix was added to the sediment, thoroughly mixed by vortex and incubated in a bath for 20 min at 56 °C. Later, reincubated for 8 min at 100 °C and centrifuged at 12,000 revolutions per minute for 3 min. The DNA was stored at − 20 °C.
Antimicrobial susceptibility testing and detection of AMR genes
The antimicrobial susceptibility testing was conducted on all enterococci isolates following the recommendations and breakpoints of the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2022). The antimicrobial agents tested were as follows (µg/disk): penicillin (10), erythromycin (15), gentamicin (120), streptomycin (300), tetracycline (30), ciprofloxacin (5), chloramphenicol (30), linezolid (10), vancomycin (30) and teicoplanin (30).
Based on the antimicrobial resistance phenotypes of all enterococci, isolates from different samples or the same sample but of different species and/or different AMR phenotypes were selected for further studies (considered as distinct isolates) (Supplementary Table S1). This collection was characterized to determine the AMR genes and genetic lineages. MDR was defined by phenotypic resistance to three or more families of antibiotics. The minimum inhibition concentration (MIC) of all isolates carrying linezolid resistance genes was tested using bioMérieux Linezolid Etest® strips (Marcy l’Étoile, France), and the results were interpreted following the EUCAST 2022 breakpoint.
The corresponding AMR genes for all antibiotics were tested by PCRs and selected according to the resistance phenotype: erythromycin (ermA, ermB, ermC, and ermT), gentamicin (aac6’-aph2″), streptomycin (str and ant6’), tetracycline (tetL, tetM, and tetK), chloramphenicol (catpC221, catpC223, catpC194, catA, fexA, and fexB), linezolid (optrA, poxtA, cfr, cfrB, and cfrD), and vancomycin (vanA and vanB). Specifically, all chloramphenicol-resistant isolates were tested for the possible presence of linezolid resistance genes and mutations in the 23S rRNA, regardless of the linezolid zone of inhibition by antibiogram. All isolates positive for linezolid resistance genes were confirmed by sequencing.
Genetic characterization
Multilocus sequence typing (MLST) was performed for E. faecalis and E. faecium isolates carrying linezolid resistance genes. The 7 housekeeping genes (gdh, gyd, pstS, gki, aroE, xpt and yqiL of E. faecalis; adK, atpA, ddl, gdh, gyd, pstS and purK of E. faecium) were amplified and sequenced, and Sequenced Types (ST) were assigned from analyses on the MLST database (https://pubmlst.org/organisms/enterococcus-faecalis). Primers and conditions of PCRs for the AMR genes tested and for MLST typing are included in Supplementary Table S2.
Data management and analyses
Data collected were verified, entered and analysed with Statistical Package for Social Sciences (SPSS) Version 26 (IBM, California, USA). Data were reported as numbers and percentages (for categorical variables). Tables and charts were plotted. Data were subjected to univariate logistics to compute Odds Ratio (OR) and chi-squared test at 95% confidence interval (95%CI) between the carriage rate of enterococci and some categorical variables (such as household densities, animal species and AMR phenotypes). A significant association was set < 0.05 probability value.
Results
Nasal enterococcal carriage rate in healthy pigs and pig-farmers
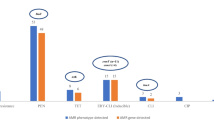
Enterococci nasal carriage was found in all the farms. In total, 51 enterococci isolates (43 from pigs, and 8 from pig-farmers) were recovered. Of the pigs’ isolates, 34, 4, 2, 2, and 1 were E. faecalis, E. faecium, E. gallinarum, E. hirae and E. casseliflavus, respectively. However, among the enterococci isolates from the pig-farmers, they were only 4 E. faecium and 4 E. faecalis isolates (Fig. 1). Of the 40 pigs studied, 29 (72.5%) were enterococci nasal carriers. Of these, 4 (40%), 9 (90%), 8 (80%) and 8 (80%) were obtained in farms-A to D, respectively (Fig. 1). Specifically, nasal carriage of E. faecalis (n = 2), E. casseliflavus (n = 1) and E. hirae (n = 1) were identified in pigs of farm-A; E. faecalis (n = 9) from pigs of farm-B; E. faecalis (n = 3), E. faecium (n = 2), E. hirae (n = 1), and E. gallinarum (n = 2) from farm-C; and E. faecalis (n = 6), E. faecalis/E. faecium co-carriage (n = 2) from farm-D (Fig. 1). Conversely, all the three farmers (100.0%) in farm-B were enterococci nasal carriers (66.7% E. faecalis and 33.3% E. faecium); 1 of the 2 farmers (50.0%) in farm-C was a nasal carrier (E. faecium); 2 of the 3 farmers of farm-D (66.7%) were nasal carriers (50.0% E. faecalis and 50.0% E. faecium), but none of the farmers in farm-A were enterococci nasal carriers (Fig. 1).
Antimicrobial resistomes of Enterococcus sp. isolates from pigs farms
In farms-A to -D, all the E. faecalis isolates from pigs and farmers were MDR (Table 1). Multiresistance was also detected in other species such as the E. casseliflavus (carrying fexA, optrA, cfrD, tetK, tetL, and ermB genes), E. hirae and E. gallinarum (carrying ermB, tetM, and ant6 genes).
None of the enterococci showed resistance to linezolid by disk diffusion; however, most of the chloramphenicol resistant isolates carried some acquired linezolid resistance genes (optrA and in some cases cfrD). Of special relevance was the detection of linezolid-resistance genes in enterococci of pigs: (a) in 33.3% of pigs tested (E. faecalis with optrA and/or cfrD; E. casseliflavus with optrA and cfrD); (b) in 10% of pig farmers (E. faecalis with optrA). The MLST of three E. faecalis isolates carrying the linezolid resistance genes from farms-A (pig) and B (pig and farmer) was performed and found to be of the genetic lineage ST330. However, the MLST of three of the linezolid resistant E. faecalis isolates from farm-D obtained from two pigs were ST330, ST474 and ST59 (Table 1). The isolates that carried linezolid resistance genes showed an MIC for linezolid in the range 8–16 µg/ml (Table 1).
Nasal enterococcal carriage rate in healthy dogs and human household members
Six out of the 27 households (22.2%) had nasal enterococci carriers. In total, 31 enterococci isolates (27 from dogs, 4 from human household members) were recovered. Of the dogs’ isolates, 18, 7 and 2 were E. faecium, E. faecalis, and E. raffinosus, respectively. However, all the isolates of humans were E. faecalis (n = 4) (Fig. 2). The nasal carriage rate of enterococci among healthy dogs and dog-owning humans were 29.4% and 4.9%, respectively.
Antimicrobial resistomes of Enterococcus sp. isolates from healthy dogs and dog-owning human household members
In one of the 6 households with enterococci carriage (household ID number 10), both humans and dogs were E. faecalis carriers and the isolates presented a similar AMR phenotype and genotype (tetM positive) (Table 2). Moreover, in another household (household ID number 18), enterococci were detected in both humans and dogs, but belonged to different species (E. faecalis in humans and E. faecium in dogs). One of the dogs analysed in this study (household ID number 18) carried linezolid-resistant E. faecalis isolates that contained the fexA, optrA, tetL, tetM, ermA, ermB, str, aac6'-aph2'', and ant6' genes, and were typed as ST585.
Antimicrobial resistomes in the Enterococcus isolates from white stork nestlings
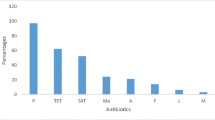
More than 70% of the 144 Enterococcus sp. from nasal and tracheal samples of stork origin studied were susceptible to all antibiotics tested (Table 3). However, 13.2% of enterococci showed tetracycline resistance, all of them of the species E. faecalis and E. faecium, and they carried the tetM and/or tetK genes (except in one E. faecium isolate); moreover, between 4 and 5% of enterococci showed erythromycin resistance (with ermB and ermA genes) and high-level aminoglycoside resistance (with aac6'-aph2'' or str genes) (Table 3). In addition, and for the first time in this animal species, an E. faecium isolate was found carrying an acquired linezolid resistance gene (poxtA), in addition to fexB gene (associated to chloramphenicol resistance); this strain belonged to the lineage ST1736 and presented an MIC for linezolid of 8 µg/ml. None of the E. casseliflavus, E. hirae, E. durans, and E. gallinarum isolates of stork origin showed resistance to the antibiotics tested (Table 3).
Comparison of AMR phenotype frequencies among E. faecalis and E. faecium
To compare the AMR frequencies of distinct E. faecalis and E. faecium isolates from dogs, pigs and storks’ nasal samples, individual chi-squared tests against every antimicrobial agent were computed. The prevalence of tetracycline, erythromycin, chloramphenicol, gentamicin, linezolid, and streptomycin resistances was significantly higher among E. faecalis of pigs than in the other two groups (p < 0.0001) (Table 4). All the enterococci carrying linezolid resistance genes were phenotypically susceptible by disc diffusion tests; however, upon LZD Etest for their MIC, all were found resistant (range: 8 to 16 μg/ml) except six isolates (all of the same animal) with an MIC of 2–3 μg/ml (Tables 1, 2 and 3).
Concerning E. faecium isolates, penicillin resistance was significantly present among isolates of dogs than in the other two groups (p < 0.05) (Table 4). However, gentamicin, erythromycin, ciprofloxacin, and streptomycin resistances were significantly higher among E. faecium of pigs than in the other two groups (Table 4). In all cases, storks’ nasal E. faecalis and E. faecium isolates had the least AMR rates compared to the dogs’ and pigs’ isolates.
Among the chloramphenicol-resistant enterococci, notably many also harbouring linezolid resistance genes (optrA, poxtA, and cfrD) were detected in 16 pigs (33.3%), 1 dog (2.9%), 1 stork (1.1%) and 1 pig-farmer (10.0%) (Fig. 3).
Risk factors associated with nasal enterococcal carriage and MDR phenotypes
After bivariate logistic analysis, nasal carriage (OR = 6.33, 95% CI: 2.29–17.42, p = 0.004) and occurrence rate of MDR phenotype (OR = 8.57, 95% CI: 2.99–24.56, p = 0.0001) were significantly associated with the species of animal (Supplementary Table S3). Although nasal enterococcal carriage in storks was double but not significantly different from that of dogs (OR = 2.4, 95% CI: 0.93–6.17, p = 0.069). Also, nasal enterococci carriage in humans was significantly associated with the species of animal contact (OR = 29.25, 95% CI: 4.36–196.07, p = 0.0005). Dog-owning households with > 1 dog & 1 human had relatively higher odds of nasal enterococci carriage than those with only 1 dog & 1 human; however, this was not statistically significant (OR = 3.75, 9% CI: 0.37–37.94, p = 0.268) (Supplementary Table S3).
Discussion
We are not aware of any previous study that simultaneously investigated the nasal enterococci communities of food-producing animals, pets and wild animals. Perhaps because they are frequent intestinal commensals, most studies focus on the GI enterococci carriage [5].
Over the last 2 decades, our research group has detected ARGs to critical antimicrobials used as the last resort chemotherapy against enterococci infections in isolates from Spain, Portugal and Tunisia (especially, conferring resistance to vancomycin and linezolid) in wild boars (vanA-carrying E. faecium), wild rodents (vanB2-carrying E. faecalis and vanA-carrying E. faecium), wild birds (vanA- and vanB2-carrying E. faecalis), chickens (vanA-carrying E. hirae), pig environment air (poxtA- and optrA-carrying E. faecium) and clinical samples (vanA/vanB E. faecalis and E. faecium, optrA- and cfrD-carrying E. faecalis) [25,26,27,28,29]. However, none of them was on nasal samples.
In all the three animal hosts studied, the nasal carriage rate was high (especially in pigs and storks). The high nasal enterococci rate detected in our study highlights their frequent association of Enterococcus spp. with the respiratory tracts of the animals. Thus, it is essential to remark that enterococci are not only found at high rates in the GI tract but also in nasal samples, as demonstrated in this study. On the other hand, healthy dogs were relatively fewer carriers of enterococci, and this might be due to host adaption differences to respiratory epithelia.
There is growing evidence that the use of chloramphenicol chemotherapy in animal husbandry can select for enterococci harbouring optrA and poxtA genes which confer resistance to the critically important antibiotic linezolid, posing a risk to human health via the food chain and contact with livestock.
In this study, the majority (over 90%) of the enterococci carrying oxazolidinone resistance genes belonged to E. faecalis or E. faecium, which are the predominant Enterococcus species in humans and animals (including pets and pigs), but also belong to the most important Gram-positive nosocomial pathogens worldwide [4]. We found a significantly higher frequency of LRE in pigs than what was reported in comparable studies on faecal samples from pigs in Switzerland (5%), Belgium (11%) and Italy (21%) [12, 13, 30]. Notably, comparative data are still scarce and variations between countries for which data are available should be interpreted with caution due to the differences in study designs, sample types and testing methodologies. Nevertheless, the present study indicates that the occurrence of chloramphenicol-resistant enterococci among our pigs are high. Worryingly, the use of antibiotics in pig farming in recent years has been very high in Spain, highlighting the need to raise awareness within the agricultural sector to mitigate the emergence and spread of linezolid-resistant enterococci in the future. Moreover, most of the enterococci in this study were associated with the presence of tetracycline resistance genes. Tetracycline is the most frequent veterinary antibiotic used for treating many swine bacterial diseases and is likely to promote the spread and persistence of LRE in pigs [31, 32]. The optrA-carrying-E. faecalis-ST330, -ST474 and -ST59 circulating in 3 of the 4 studied farms have been previously reported in human and many animal hosts such as macaques, pigs, chickens, poultry meat, and vultures [13, 30, 33,34,35,36,37,38,39,40]. These optrA-positive lineages appear to be non-host specific.
The detection of LRE in pig farmers and a dog indicate potential risk of transmission to other humans and animals outside the pigs-farm environment and dog-owning households, respectively. These put together with the several optrA-positive E. faecalis isolates reported in dogs fed with raw meat/vegetables in China [41] underscore relevance of the ‘One-Health’ approach for investigating LRE, as they can be shared by animals, humans and environnment. However, the direction of transfer is often difficult to prove, especially as none of the humans in contact with the dogs were carriers of LRE. Currently, the knowledge of the LRE prevalence in companion animals is limited and therefore a joint approach to monitor the emergence and dissemination of resistance mechanisms of public health importance are needed. The MDR E. faecalis-ST585 isolate detected in a dog in our study was similar to previously reported LR-E. faecalis isolates from Spanish hospitals [42]. Moreover, this is the first description of ST585 carrying the optrA gene in dogs from Spain.
Plasmid-encoded optrA and poxtA in E. durans and E. hirae were previously reported in pigs in Switzerland, as well as poxtA-carrying E. hirae from China and Italy [13, 29, 31, 43] and optrA-carrying E. gallinarum from a fattening pig in Belgium [12] were recently identified. Also, a cfrD-carrying E. casseliflavus strain has recently been reported from pigs’ manure in Italy [44] and optrA/cfr-carrying E. casseliflavus from a faecal swab of a pig in China [45]. To the best of our knowledge, the detection of E. casseliflavus carrying optrA and cfrD in a pig in our study is the first report. These put together suggest that pigs could be potential reservoirs for the dissemination and persistence of E. casseliflavus carrying various linezolid transferable resistance genes. As E. casseliflavus only occasionally causes opportunistic infections in humans [4], the presence of linezolid resistance genes in this species from pigs may not pose a direct threat to human health but could play an important role in transferring this resistance mechanism. It is worth mentioning that none of the isolates had mutation in their 23S rDNA.
Concerning the stork’s E. faecium-ST1736 carrying poxtA in our study, migratory birds may be an important link in the spread of LRE. This isolate was obtained from a nestling that was feed food foraged by its parents in the landfills; so, the exposure could be from human household residues, landfill discarded wastewater treatment plant slurry or animal remains. This is the first time that LR-E. faecium ST1736 has been reported in storks. The detection of linezolid resistance genes is highly relevant since these genes could be in plasmids and be transmitted to clinical settings, production animals or the environment.
It is of interest to remark that all the linezolid resistant enterococci were recovered in the ChromAgar LIN agar plates in which isolates were grown as green colonies. Nevertheless, linezolid susceptible isolates were also recovered in this media, as also indicated by other authors [24].
In storks, a vast majority of the Enterococcus species were susceptible to all the antibiotics tested. This difference may reflect the level of selection pressure, particularly due to the extensive use of antibiotics in pig-farming as compared to antibiotic chemotherapy in dogs and humans [46]. Although vancomycin-resistant enterococci (VRE) are considered high-priority pathogens of great public health concern resistance [47], none of the isolates carried the vanA and vanB genes. Notably, the detected AMR genes in E. faecium or faecalis isolates from storks were all from landfill-associated colonies (except one). Most likely, the individuals were fed landfill foraged food by their parents.
Both acquired and intrinsic resistance properties drastically reduce the options for antimicrobial therapy. Bearing this in mind, the performance of antimicrobial susceptibility tests prior to the start of antimicrobial therapy is of particular significance to guide the application of antimicrobial agents in pigs-farming and canine medicine.
Conclusion
Enterococcus casseliflavus carrying optrA and cfrD is described for the first time in pigs. This put together with the occurrence of cfrD and optrA in E. faecalis-ST585 and -ST330, poxtA in E. faecium-ST1736 in healthy dogs, pigs and storks emphasizes the potential risk to human health through the dissemination of LRE in the food chain and companion and wild animals. The poxtA gene is described for the first time in an E. faecium from a migratory bird that could facilitate its spread to other ecosystems. Foraging of the parents of this stork in landfills could explain the acquisition of this multidrug-resistant strain. The presence of linezolid resistance genes with potential of horizontal transfer may go unnoticed by disc diffusion phenotypic tests, unless they are detected by MIC determination. Nevertheless, the selective inclusion of chloramphenicol resistance phenotype as marker to screen for linezolid resistance genes in enterococci may be advantageous for their detection at a molecular level. Our results showed that the nasal cavity of pigs, dogs and the trachea of storks may represent an important source of LRE, possibly contributing to animal-to-human transmission and transmission to the environment or food by these colonized animals and people.
Data Availability
The data generated from this study can be available on request through the corresponding author (Carmen Torres).
References
Ferchichi M, Sebei K, Boukerb AM, Karray-Bouraoui N, Chevalier S, Feuilloley MGJ et al (2021) Enterococcus spp.: is it a bad choice for a good use-a conundrum to solve? Microorganisms 9(11):2222. https://doi.org/10.3390/microorganisms9112222
Dubin K, Pamer EG (2014) Enterococci and their interactions with the intestinal microbiome. Microbiol Spectr 5(6):https://doi.org/10.1128/microbiolspec.BAD-0014-2016. https://doi.org/10.1128/microbiolspec.BAD-0014-2016
Cattoir V (2022) The multifaceted lifestyle of enterococci: genetic diversity, ecology and risks for public health. Curr Opin Microbiol 65:73–80. https://doi.org/10.1016/j.mib.2021.10.013
Zaheer R, Cook SR, Barbieri R, Goji N, Cameron A, Petkau A et al (2020) Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci Rep 10(1):3937. https://doi.org/10.1038/s41598-020-61002-5
Torres C, Alonso CA, Ruiz-Ripa L, León-Sampedro R, Del Campo R, Coque TM (2018) Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol Spectr 6(4). https://doi.org/10.1128/microbiolspec.ARBA-0032-2018
Mupfunya CR, Qekwana DN, Naidoo V (2021) Antimicrobial use practices and resistance in indicator bacteria in communal cattle in the Mnisi community, Mpumalanga, South Africa. Vet Med Sci 7(1):112–121. https://doi.org/10.1002/vms3.334
Vrancianu CO, Popa LI, Bleotu C, Chifiriuc MC (2020) Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front Microbiol 6(11):761. https://doi.org/10.3389/fmicb.2020.00761
Woolhouse M, Ward M, Van Bunnik B, Farrar J (2015) Antimicrobial resistance in humans, livestock and the wider environment. Phil Trans R Soc B: Biol Sci 370(1670):20140083. https://doi.org/10.1098/rstb.2014.0083
European Commission (2022) Regulation (EU) 2019/6 of the European Parliament and of the Council of 11 December 2018 on veterinary medicinal products and repealing Directive 2001/82/EC. https://eur-lex.europa.eu/eli/reg/2019/6/oj Last Accessed 3rd October 2022
Daniel DS, Lee SM, Dykes GA, Rahman S (2015) Public health risks of multiple-drug-resistant Enterococcus spp. in Southeast Asia. Appl Environ Microbiol 81(18):6090–6097. https://doi.org/10.1128/AEM.01741-15
Lees P, Pelligand L, Giraud E, Toutain PL (2021) A history of antimicrobial drugs in animals: evolution and revolution. J Vet Pharmacol Ther 44(2):137–171. https://doi.org/10.1111/jvp.12895
Timmermans M, Bogaerts B, Vanneste K, De Keersmaecker SCJ, Roosens NHC, Kowalewicz C et al (2021) Large diversity of linezolid-resistant isolates discovered in food-producing animals through linezolid selective monitoring in Belgium in 2019. J Antimicrob Chemother 77(1):49–57. https://doi.org/10.1093/jac/dkab376
Fioriti S, Morroni G, Coccitto SN, Brenciani A, Antonelli A, Di Pilato V et al (2020) Detection of oxazolidinone resistance genes and characterization of genetic environments in Enterococci of swine origin, Italy. Microorganisms 8(12):2021. https://doi.org/10.3390/microorganisms8122021
Elghaieb H, Tedim AP, Abbassi MS, Novais C, Duarte B, Hassen A et al (2020) From farm to fork: identical clones and Tn6674-like elements in linezolid-resistant Enterococcus faecalis from food-producing animals and retail meat. J Antimicrob Chemother 75(1):30–35. https://doi.org/10.1093/jac/dkz419
Gouliouris T, Raven KE, Ludden C, Blane B, Corander J, Horner CS et al (2018) Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. mBio 9(6):e01780-18. https://doi.org/10.1128/mBio.01780-18
Willems RJ, Hanage WP, Bessen DE, Feil EJ (2011) Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol Rev 35(5):872–900. https://doi.org/10.1111/j.1574-6976.2011.00284.x
van den Bogaard AE, Willems R, London N, Top J, Stobberingh EE (2002) Antibiotic resistance of faecal enterococci in poultry, poultry farmers and poultry slaughterers. J Antimicrob Chemother 49(3):497–505. https://doi.org/10.1093/jac/49.3.497
Jung WK, Shin S, Park YK, Noh SM, Shin SR, Yoo HS et al (2020) Distribution and antimicrobial resistance profiles of bacterial species in stray dogs, hospital-admitted dogs, and veterinary staff in South Korea. Prev Vet Med 184:105151. https://doi.org/10.1016/j.prevetmed.2020.105151
Ghosh A, Dowd SE, Zurek L (2011) Dogs leaving the ICU carry a very large multi-drug resistant enterococcal population with capacity for biofilm formation and horizontal gene transfer. PLoS One 6(7):e22451. https://doi.org/10.1371/journal.pone.0022451
Feßler AT, Scholtzek AD, Schug AR, Kohn B, Weingart C, Hanke D, Schink AK et al (2022) Antimicrobial and biocide resistance among canine and feline Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii isolates from diagnostic submissions. Antibiotics (Basel) 11(2):152. https://doi.org/10.3390/antibiotics11020152
Sterneberg-van der Maaten T, Turner D, Van Tilburg J, Vaarten J (2016) Benefits and risks for people and livestock of keeping companion animals: searching for a healthy balance. J Comp Pathol 155(1 Suppl 1):S8–S17. https://doi.org/10.1016/j.jcpa.2015.06.007
Vittecoq M, Godreuil S, Prugnolle F, Durand P, Brazier L, Renaud N et al (2016) Antimicrobial resistance in wildlife. J Appl Ecol 53(2):519–529
Abdullahi IN, Juárez-Fernández G, Höfle U, Cardona-Cabrera T, Mínguez D, Pineda-Pampliega J, Lozano C, Zarazaga M, Torres C (2023). Nasotracheal microbiota of white storks with different foraging habits, in Spain. EcoHealth (in press)
Girlich D, Mihaila L, Cattoir V, Laurent F, Begasse C, David F, Metro CA, Dortet L (2022) Evaluation of CHROMagar™ LIN-R for the screening of linezolid resistant Staphylococci from positive blood cultures and nasal swab screening samples. Antibiotics (Basel) 11(3):313. https://doi.org/10.3390/antibiotics11030313
Robredo B, Singh KV, Baquero F, Murray BE, Torres C (1999) From vanA Enterococcus hirae to vanA Enterococcus faecium: a study of feed supplementation with avoparcin and tylosin in young chickens. Antimicrob Agents Chemother 43(5):1137–1143. https://doi.org/10.1128/AAC.43.5.1137
Poeta P, Costa D, Igrejas G, Rojo-Bezares B, Sáenz Y, Zarazaga M et al (2007) Characterization of vanA-containing Enterococcus faecium isolates carrying Tn5397-like and Tn916/Tn1545-like transposons in wild boars (Sus Scrofa). Microb Drug Resist 13(3):151–6. https://doi.org/10.1089/mdr.2007.759
Ben Yahia H, Chairat S, Hamdi N, Gharsa H, Ben Sallem R, Ceballos S et al (2018) Antimicrobial resistance and genetic lineages of faecal enterococci of wild birds: emergence of vanA and vanB2 harbouring Enterococcus faecalis. Int J Antimicrob Agents 52(6):936–941. https://doi.org/10.1016/j.ijantimicag.2018.05.005
Ruiz-Ripa L, Feßler AT, Hanke D, Eichhorn I, Azcona-Gutiérrez JM, Pérez-Moreno MO et al (2020) Mechanisms of linezolid resistance among Enterococci of clinical origin in Spain-detection of optrA- and cfr(D)-carrying E.faecalis. Microorganisms 8(8):1155. https://doi.org/10.3390/microorganisms8081155
Ruiz-Ripa L, Feßler AT, Hanke D, Sanz S, Olarte C, Eichhorn I et al (2020) Detection of poxtA- and optrA-carrying E. faecium isolates in air samples of a Spanish swine farm. J Glob Antimicrob Resist 22:28–31. https://doi.org/10.1016/j.jgar.2019
Nüesch-Inderbinen M, Haussmann A, Treier A, Zurfluh K, Biggel M, Stephan R (2022) Fattening pigs are a reservoir of florfenicol-resistant Enterococci harboring oxazolidinone resistance genes. J Food Prot 85(5):740–746. https://doi.org/10.4315/JFP-21-431
Schwarz S, Zhang W, Du XD, Krüger H, Feßler AT, Ma S et al (2021) Mobile oxazolidinone resistance genes in gram-positive and gram-negative bacteria. Clin Microbiol Rev 34(3):e0018820. https://doi.org/10.1128/CMR.00188-20
Swiss Veterinary Society (SVS) (2018) Umsichtiger Einsatz von Antibiotika bei Rindern und Schweinen: Therapieleitfaden fur Tierarztinnen und Tierarzte. Available at: https://www.blv.admin.ch/dam/blv/de/dokumente/tiere/tierkrankheiten-und. Accessed 6 Oct 2022
Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z et al (2015) A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70(8):2182–2190. https://doi.org/10.1093/jac/dkv116
McHugh MP, Parcell BJ, Pettigrew KA, Toner G, Khatamzas E, El Sakka N et al (2022) Presence of optrA -mediated linezolid resistance in multiple lineages and plasmids of Enterococcus faecalis revealed by long read sequencing. Microbiology (Reading) 168(2):001137. https://doi.org/10.1099/mic.0.001137
Freitas AAR, Souza SDSR, Faria AR, Planet PJ, Merquior VLC et al (2022) Draft genome sequences of two commensal Enterococcus faecalis strains isolated from American black vultures (Coragyps atratus) in Brazil. Microbiol Resour Announc 11(8):e0005722. https://doi.org/10.1128/mra.00057-22
Cavaco LM, Bernal JF, Zankari E, Léon M, Hendriksen RS, Perez-Gutierrez E et al (2017) Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother 72(3):678–683. https://doi.org/10.1093/jac/dkw490
Woods SE, Lieberman MT, Lebreton F, Trowel E, de la Fuente-Núñez C, Dzink-Fox J et al (2017) Characterization of multi-drug resistant Enterococcus faecalis isolated from cephalic recording chambers in research macaques (Macaca spp.). PLoS One 12(1):e0169293. https://doi.org/10.1371/journal.pone.0169293
Almeida LM, Gaca A, Bispo PM, Lebreton F, Saavedra JT, Silva RA et al (2020) Coexistence of the oxazolidinone resistance-associated genes cfr and optrA in Enterococcus faecalis from a healthy piglet in Brazil. Front Public Health 24(8):518. https://doi.org/10.3389/fpubh.2020.00518
Roy S, Aung MS, Paul SK, Ahmed S, Haque N, Khan ER et al (2020) Drug resistance determinants in clinical isolates of Enterococcus faecalis in Bangladesh: identification of oxazolidinone resistance gene optrA in ST59 and ST902 lineages. Microorganisms 8(8):1240. https://doi.org/10.3390/microorganisms8081240
Càmara J, Camoez M, Tubau F, Pujol M, Ayats J, Ardanuy C et al (2019) Detection of the novel optrA gene among linezolid-resistant Enterococci in Barcelona, Spain. Microb Drug Resist 25(1):87–93. https://doi.org/10.1089/mdr.2018.0028
Wu Y, Fan R, Wang Y, Lei L, Feßler AT, Wang Z et al (2019) Analysis of combined resistance to oxazolidinones and phenicols among bacteria from dogs fed with raw meat/vegetables and the respective food items. Sci Rep 9(1):15500. https://doi.org/10.1038/s41598-019-51918-y
Moure Z, Lara N, Marín M, Sola-Campoy PJ, Bautista V, Gómez-Bertomeu F et al (2020) Interregional spread in Spain of linezolid-resistant Enterococcus spp. isolates carrying the optrA and poxtA genes. Int J Antimicrob Agents 55(6):105977. https://doi.org/10.1016/j.ijantimicag.2020.105977
He T, Shen Y, Schwarz S, Cai J, Lv Y, Li J et al (2016) Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J Antimicrob Chemother 71(6):1466–1473. https://doi.org/10.1093/jac/dkw016
Cinthi M, Coccitto SN, Fioriti S, Morroni G, Simoni S, Vignaroli C et al (2022) Occurrence of a plasmid co-carrying cfr(D) and poxtA2 linezolid resistance genes in Enterococcus faecalis and Enterococcus casseliflavus from porcine manure, Italy. J Antimicrob Chemother 77(3):598–603. https://doi.org/10.1093/jac/dkab456
Lei CW, Chen X, Liu SY, Li TY, Chen Y, Wang HN (2021) Clonal spread and horizontal transfer mediate dissemination of phenicol-oxazolidinone-tetracycline resistance gene poxtA in enterococci isolates from a swine farm in China. Vet Microbiol 262:109219. https://doi.org/10.1016/j.vetmic.2021.109219
Benjamin JK, Bruce AH, Lance BP (2017) Food-animal production and the spread of antibiotic resistance: the role of ecology. Front Ecol Environ 15(6):309–318. https://doi.org/10.1002/fee.1505
European Centre for Disease Prevention and Control (2017) ECDC tool for the prioritization of infectious disease threats – handbook and manual. ECDC, Stockholm
Acknowledgements
Parts of this study (related to E. faecium of storks) was presented as poster in the XXV Congress (2022) of the Spanish Society of Infectious Diseases and Clinical Microbiology. We acknowledge the support of the veterinarians and other people that participated in the sampling process. We are very grateful to Dr H. Cruz Ramos and CHROMagarTM (Paris, France) for providing CHROMAgar LIN™ used in this study.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by the project PID2019-106158RB-I00 of the MCIN/ AEI /10.13039/501100011033 of Spain and project SBPLY/19/180501/000325 of the regional government of Castilla – La Mancha co-financed by the European Union’s funds for regional development (Feder). Also, it received funding from the European Union’s H2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 801586.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.T., I.N.A.; methodology: C.T., I.N.A.; sampling: I.N.A., U.H., C. S.; experimental work: I.N.A., G.J.F., B.M-C, S.R., A.M., S.M.A., P.E; validation: C.T., I.N.A., C.L., M.Z., U.H.; formal analysis: C.T., I.N.A., C.L., M.Z., U.H.; software analysis: I.N.A; data curation: C.T., I.N.A., C.L., M.Z., U.H.; writing—original draft preparation: I.N.A., C.T., C.L., M.Z.; writing—review and editing: I.N.A., C.T., C.L., M.Z., G. J.F., B.M.C, S.R., A.M., S.M.A., P.E.; project administration: C.T., M.Z.; funding acquisition: C.T., M.Z., I.N.A; U.H. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdullahi, I.N., Lozano, C., Juárez-Fernández, G. et al. Nasotracheal enterococcal carriage and resistomes: detection of optrA-, poxtA- and cfrD-carrying strains in migratory birds, livestock, pets, and in-contact humans in Spain. Eur J Clin Microbiol Infect Dis 42, 569–581 (2023). https://doi.org/10.1007/s10096-023-04579-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-023-04579-9