Abstract
PCR detection of Helicobacter pylori infection in gastric biopsies allows the detection of this bacterium and the mutations associated with macrolide resistance. The aim of this study was to evaluate the performance of RIDA®GENE H. pylori PCR (r-Biopharm) on the ELITe InGenius System (Elitech). Two hundred gastric biopsies were obtained. These biopsies were ground in nutrient broth. Two hundred microliters of this suspension was treated with proteinase K, and then, 200 µL was transferred to an ELITe InGenius sample tube and tested using RIDA®GENE H. pylori PCR reagents. In-house H. pylori PCR was used as a reference. The sensitivity of RIDA®GENE H. pylori PCR with ELITe InGenius was 100%, the specificity was 98% (95% confidence interval (CI), 95.3–100%), the PPV was 98% (95% CI, 95.3–100%), and the NPV was 100% for the detection of H. pylori. All of these parameters were 100% for the categorization of macrolide resistance. The adaptation of RIDA®GENE H. pylori PCR reagents on the ELITe InGenius System was successful. This PCR is easy to use on this system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Of all the techniques available for the diagnosis of Helicobacter pylori infection in gastric biopsies, PCR is now widely used in microbiology laboratories. It is more sensitive than culture [1] and provides information on the presence of infection and macrolide resistance mutations within hours. Many commercial kits have been developed, and their performances have been evaluated. Our laboratory has participated in the evaluation of some of them [2,3,4].
We previously demonstrated that RIDA®GENE H. pylori PCR (r-Biopharm, Courtaboeuf, France) [2] could be adapted to the BD MAX™ device commercialized by Becton Dickinson (Le Pont de Claix, France) [5, 6]. This kind of system has the advantage of full automation of DNA extraction, PCR amplification, and analysis of the results. The aim of our study was to evaluate RIDA®GENE H. pylori PCR on an equivalent system, the ELITe InGenius from the Elitech Group (Puteaux, France). This evaluation was performed retrospectively using 200 frozen gastric biopsies with known H. pylori status and macrolide sensitivity.
Material and methods
Study design
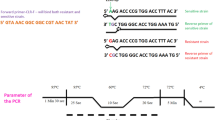
The procedure for the ELITe InGenius System was optimized for the isolation of DNA from 200 µL samples. ELITe InGenius contains a combination of lytic and extraction reagents designed to perform cell lysis and DNA extraction. Following cell lysis, the released DNA is captured by magnetic affinity beads. The beads with the bound DNA were washed, and then, the DNA was eluted using 100 μL of elution buffer. The eluted DNA may be used for applications on the ELITe InGenius. The PCR amplification parameters used on the ELITe InGenius were denaturation at 95 °C for 60 s (1 cycle) followed by 95 °C for 15 s and 60 °C for 30 s of amplification and detection (45 cycles). Channels 1 (Ct threshold 50), 2 (Ct threshold 50), and 5 (Ct threshold 200) were used for H. pylori, internal control (IC), and clarithromycin resistance detection, respectively.
Two hundred gastric biopsies obtained from the National Reference Center for Campylobacters and Helicobacters (NRCCH) (www.cnrch.fr) received at the NRCCH during April and July 2021 were included (Suppl Table 1). They were acquired from 105 women and 95 men (sex ratio 0.47) with an average age of 49 years ± 16.9. On receipt, according to a routine protocol, these biopsies were previously ground in 1 mL of nutrient broth and then stored at −80 °C. Part of this suspension was treated with 20 μL of proteinase K (pK) (Roche Diagnostic, Meylan, France) + 180 μL of ATL Qiagen buffer at 56 °C for 3 h, and 200 µL was used for the test on the ELITe InGenius. The same digestion protocol was used on ground biopsies before DNA extraction on a MagNA Pure 96 system (Roche Diagnostics) and PCR on Eurogentec strips (Liège, Belgium) using an LC480 (Roche Diagnostics, Meylan, France) as previously described [7]. The culture was performed in parallel according to internal laboratory procedures [8].
Limits of detection
The reference H. pylori strain CCUG17874 (susceptible to clarithromycin) and a clinical strain from a routine procedure (isolated from the gastric biopsy of a 63-year-old man) resistant to clarithromycin with an A2142 or A2143G mutation were grown on homemade Pylori agar [8] and then used to evaluate the limit of detection of RIDA®GENE PCR on the ELITe InGenius. Both strains were suspended at 1.5 McF (approximately 1.4×108 CFU/mL) in nutrient Brucella broth, and then, serial dilutions from 10−4 to 10−7 were generated. Two hundred microliters of each dilution was used for extraction and PCR analysis on the ELITe InGenius.
Reference used
H. pylori PCR from NRCCH was used as a reference [7]. In the event of a discrepancy, each biopsy was tested a second time on the ELITe InGenius, and PCR on the DNA extracted by the NRCCH with the MagNA Pure 96 extractor on a CFX96 system (Bio-Rad, Les Ulis, France) using RIDA®GENE H. pylori reagents was performed as previously described [2]. In the event of an unresolved discrepancy, clinical information was considered if available.
Results
Of the 200 biopsies, 100 were expected to be negative for H. pylori. In all, 98 biopsies were negative on the ELITe InGenius using RIDA®GENE H. pylori reagents (Table 1) (Suppl Table 1). Two biopsies were positive for H. pylori (biopsies 159 and 189, with Ct values of 35 and 38.4, respectively) (Suppl Table 1). The amplification curves of these two samples were similar to those obtained on the other H. pylori positive samples. These two cases were detected as positive again on a second passage on the ELITe InGenius. The 2 DNA samples extracted at the NRCCH for these 2 cases tested negative using RIDA®GENE H. pylori reagents on a CFX96 system (data not shown). No history or suspicion of H. pylori infection was available for these other discordant cases, and thus, they were considered false-positives (Table 1) (Table 2).
The 100 biopsies expected to be positive for H. pylori according to the NRCCH results were also positive on the ELITe InGenius using RIDA®GENE H. pylori reagents. The mean Ct value for H. pylori detection for these 100 biopsies was 23.3 Ct ± 2.5. The performance of RIDA®GENE H. pylori PCR on an ELITe InGenius for H. pylori detection was therefore determined to be as follows: sensitivity 100%, specificity 98% (95% confidence interval (CI), 95.3.21–100%), negative predictive value (NPV) 100%, and positive predictive value (PPV) 98% (95% CI, 95.3–100%).
The limit of detection of H. pylori (see “Material and methods”) on the ELITe InGenius was approximately 65 CFU (data not shown).
Regarding the detection on the ELITe InGenius of the 23S rDNA genotype associated with sensitivity to macrolides, a population sensitive to macrolides (WT genotype) was expected for 65 biopsies: these samples were perfectly detected and categorized on the ELITe InGenius. For 30 biopsies, H. pylori with an A2142G or A2143G mutation was expected: they were perfectly detected and categorized on the ELITe InGenius. For 5 biopsies, a mixed WT + A2142G or A2143G-mutated population was expected (Table 1) (Table 2) (Suppl Table 1), and they were perfectly categorized.
The mean Ct value for detecting clarithromycin resistance was 26.7 ± 2.4. The detection performance of the 23S rDNA genotype on the ELITe InGenius was therefore perfect, with 100% sensitivity, specificity, NPV, and PPV.
Discussion
The objective of our study was to evaluate RIDA®GENE H. pylori PCR on an ELITe InGenius using tissue from gastric biopsies. The performance validation was conducted by a retrospective study of 200 gastric biopsies. Our results showed that this adaptation is possible, and the results obtained were excellent.
The advantage of automated RIDA®GENE H. pylori PCR on an ELITe InGenius is to provide the possibility for clinical laboratories equipped with this system to detect not only H. pylori but also the mutations associated with macrolide resistance. Interpretation of the results, which is possible on the ELITe InGenius, was easy. We did not have to adjust the detection of the IC, which is an essential parameter used to interpret the results, particularly for negative samples. IC amplification can be inhibited if H. pylori is detected, but this does not interfere with interpretation. If only macrolide resistance is detected, i.e., in the absence of H. pylori detection, the test, as indicated by r-Biopharm, should be interpreted as negative. For the 100 biopsies expected to be negative according to the in-house PCR, this situation occurred only once (biopsy 142) (Suppl Table 1).
The discrepancies in this study, compared with our real-time PCR method used as a reference, were observed only in 2 samples. Two false-positive results were indeed detected for which histology reports were not available. RIDA®GENE H. pylori PCR on an ELITe InGenius correctly detects the mutations associated with macrolide resistance (point mutations at two nucleotide positions, 2142 (A2142G and A2142C) and 2143 (A2143G) [1]), even for biopsies containing a mixture of susceptible and resistant populations. However, it would be interesting to verify these results in a larger number of biopsies of this type. The A2142G or A2143G mutations are the most frequent in France, and in Europe, the A2142C mutation remains anecdotal [9, 7]. Finally, A2142T and A2143C mutations are rarely found in France (<0.1% of all macrolide-resistant strains in France (2017–2021 personal data)) [10]. Samples positive for these mutations (one sample each) were tested on the ELITe InGenius; as expected, H. pylori-only was detected (data not shown).
In conclusion, the performance of RIDA®GENE H. pylori PCR on the ELITe InGenius was excellent. The performance was similar to that published previously by our team for these same reagents but with a different DNA extraction technique and an independent real-time PCR device, the CFX96 [2] or the BD Max system [6]. This adaptation in such apparatuses allows the automation of DNA extraction, PCR amplification, and automatic interpretation of results in a single machine. The advantage of the ELITe InGenius System is to be able to recover DNA extracted by the system for controls or further analysis if needed, whereas this is not possible on the BD MAX™ where the DNA at the end of the run is mixed with the reaction mix. The capacity of the BD MAX™ is 24 samples against only 12 for the ELITe InGenius. The choice of the machine will depend on the volume of activity of each laboratory wishing to set up this PCR. The reading and interpretation of the amplification curves and the results obtained is easier on BD MAX™ than on ELITe InGenius. On the other hand, the launching of an analysis run on ELITe InGenius is guided step by step by the machine whereas it is less intuitive on BD MAX™. The number of reagents needed on BD MAX™ is however smaller than on ELITe InGenius. In the end, each system has its advantages and disadvantages which can be assessed by future users according to their working environment. Their use is very simple, and the obtained performances are excellent.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable
References
Megraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev [Internet] 2007;20:280–322. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17428887
Bénéjat L, Ducournau A, Lehours P, Mégraud F (2018) Real-time PCR for Helicobacter pylori diagnosis The best tools available. Helicobacter 23:12512. https://doi.org/10.1111/hel.12512
Jehanne Q, Bénéjat L, Mégraud F, Bessède E, Lehours P (2020) Evaluation of the AllplexTM H pylori and ClariR PCR Assay for Helicobacter pylori detection on gastric biopsies. Helicobacter 25:12702. https://doi.org/10.1111/hel.12702
Hays C, Delerue T, Lamarque D, Burucoa C, Collobert G, Billöet A et al (2019) Molecular diagnosis of Helicobacter pylori infection in gastric biopsies: Evaluation of the Amplidiag® H pylori + ClariR assay. Helicobacter 24:12560. https://doi.org/10.1111/hel.12560
Berenger BM, Chui L, Ferrato C, Lloyd T, Li V, Pillai DR. Performance of four commercial real-time PCR assays for the detection of bacterial enteric pathogens in clinical samples. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021 23;S1201-9712(21)00831-6. https://doi.org/10.1016/j.ijid.2021.10.035
Bénéjat L, Giese A, Lescaudron Z, Bonnac J, Ducournau A, Bessède E et al (2022) Automation of RIDA®GENE Helicobacter pylori PCR on the BD MAXTM System. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 41:875–9. https://doi.org/10.1007/s10096-022-04444-1
Bénéjat L, Ducournau A, Domingues-Martins C, Lecoeur M, Blosse A, Mégraud F et al (2021) Adaptation of an in-house PCR for the detection of Helicobacter pylori and the mutations associated with macrolide resistance into ready-to-use PCR microwell strips. Helicobacter 26:12855. https://doi.org/10.1111/hel.12855
Lehours P, Mégraud F (2021) Culture-Based Antimicrobial Susceptibility Testing for Helicobacter pylori. Methods Mol. Biol. Clifton NJ 2283:45–50. https://doi.org/10.1007/978-1-0716-1302-3_6
Lauener FN, Imkamp F, Lehours P, Buissonnière A, Benejat L, Zbinden R et al (2019) Genetic Determinants and Prediction of Antibiotic Resistance Phenotypes in Helicobacter pylori. J. Clin. Med. 7:8. https://doi.org/10.3390/jcm8010053
Ducournau A, Bénéjat L, Sifré E, Bessède E, Lehours P, Mégraud F (2016) Helicobacter pylori resistance to antibiotics in 2014 in France detected by phenotypic and genotypic methods. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 22:715–8. https://doi.org/10.1016/j.cmi.2016.06.003
Acknowledgments
The authors want to thank all the clinicians and laboratories who submitted samples to our reference center for H. pylori diagnosis. The current manuscript was edited for proper English language using American Journal Experts services (verification code 8A7A-7D3E-9C19-29DC-010D).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection and analysis were performed by Bénejat Lucie, Astrid Ducournau, Chloé Domingues-Martins, and Philippe Lehours. The first draft of the manuscript was written by Bénejat Lucie and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.”
Corresponding author
Ethics declarations
Ethics approval
All diagnostic methods were performed retrospectively. All patients were investigated in a hospital and private clinical setting according to good clinical practices. In this routine process, consent for the endoscopic procedure is always provided in writing and maintained in the patient’s medical record.
Consent to participate
No informed consent for using human gastric DNA was requested from the patients.
Consent for publication
Not applicable
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bénéjat, L., Ducournau, A., Martins, C.D. et al. RIDA®GENE Helicobacter pylori PCR on the ELITe InGenius System. Eur J Clin Microbiol Infect Dis 42, 593–596 (2023). https://doi.org/10.1007/s10096-023-04563-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-023-04563-3