Abstract
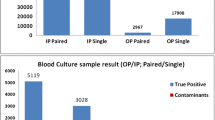
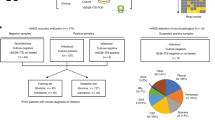
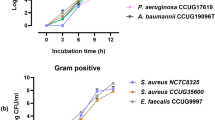
Blood cultures detected as positive by the automated system but negative by microscopy and subculture are considered as “false-positives.” Several causes have been identified, including hyperleukocytosis or the presence of fastidious bacteria, but as many cases remain unexplained we aimed to investigate the false positives occurring in our laboratory. We retrospectively collected data on blood cultures received over a period of 12 months to determine factors associated with the false-positive vials. We then prospectively validated our findings on the false-positive results occurring over a 3.5-month period. We finally applied scanning electron microscopy (SEM) on 63 false positives and molecular approaches on a subset of them. In the retrospective study, 154 (85%) of the 181 false-positive identified were positive following less than 4 h of incubation and were considered as “early false-positives.” By performing ROC curves on these early false positives, we demonstrate that the absolute number of leukocytes is in fact the most discriminating factor of early false positivity (p < 0.001). This phenomenon can be the consequence of either a high blood culture volume (p < 0.001) or hyperleukocytosis (p < 0.001). In the prospective study, the use of a threshold of 219 million of leukocytes per vial enabled the identification of 97% of the early false positives. Finally, SEM and specific qPCR enabled three additional identifications while 16S rRNA/nanopore sequencing enabled the detection of Helicobacter cinaedi bacteremia and a polymicrobial infection. A high absolute number of leukocytes in blood cultures explains most false positives, thereby making it possible to target additional microbiological investigations.



Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in Supplementary Material.
Code availability
Not applicable.
References
Miller JM, Binnicker MJ, Campbell S, Carroll KC, Chapin KC, Gilligan PH et al (2018) A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis 67(6):e1-94
Dubourg G, Raoult D (2016) Emerging methodologies for pathogen identification in positive blood culture testing. Expert Rev Mol Diagn 16(1):97–111
Emeraud C, Yilmaz S, Fortineau N, Cuzon G, Dortet L (2021) Quality indicators for blood culture: 1 year of monitoring with BacT/Alert Virtuo at a French hospital. J Med Microbiol 70(3):001300. https://doi.org/10.1099/jmm.0.001300
Ebihara Y, Kobayashi K, Watanabe N, Taji Y, Maeda T, Takahashi N et al (2019) False-positive blood culture results in patients with hematologic malignancies. J Infect Chemother 25(5):404–406
Karakonstantis S, Manika I, Vakonaki M, Boula A (2018) False-positive blood cultures in acute leukemia: an underrecognized finding. Case Rep Med 2018:7090931
Turan D, Kuruoğlu T, Gümüş D, Kalaycı F, Şerefhanoğlu K (2018) Evaluation of factors that may cause false positive growth signals in blood cultures-as the word « factors » will include both microbial and patients as well as others. Int J Clin Med Microbiol 3:137. https://www.graphyonline.com/archives/IJCMM/2018/IJCMM-137/. Accessed 13 May 2021
Qian Q, Tang YW, Kolbert CP, Torgerson CA, Hughes JG, Vetter EA et al (2001) Direct identification of bacteria from positive blood cultures by amplification and sequencing of the 16S rRNA gene: evaluation of BACTEC 9240 instrument true-positive and false-positive results. J Clin Microbiol 39(10):3578–3582
Van Laer F, Mermans I, Jansens H (2016) Flacons d’hémoculture faussement positifs et sur-remplissage. Noso-info http://www.nosoinfo.be/nosoinfos/flacons-dhemoculture-faussement-positifs-et-sur-remplissage/. Accessed 13 May 2021
Pitsch A, Ergani A, Vasse M, Farfour E (2019) False-positive of blood culture instrument: leukocytosis, overfilled vials or defective position? Ann Biol Clin (Paris) 177(6):665–667
de Vries JJC, van Assen S, Mulder AB, Kampinga GA (2007) Positive blood culture with Plasmodium falciparum: case report. Am J Trop Med Hyg Juin 76(6):1098–1099
Lee S, Kim S (2019) Accuracy of BacT/Alert Virtuo for measuring blood volume for blood culture. Ann Lab Med 39(6):590–592
Haddad G, Bellali S, Takakura T, Fontanini A, Ominami Y, Bou Khalil J et al (2021) Scanning electron microscope: a new potential tool to replace gram staining for microbe identification in blood cultures. Microorganisms 9(6):1170
Morel AS, Dubourg G, Prudent E, Edouard S, Gouriet F, Casalta JP et al (2015) Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis 34(3):561–570
Funding
This work was supported by the French Government under the “Investments for the Future” program managed by the National Agency for Research (ANR), Méditerranée-Infection 10–71 IAHU-03. populations of false-positive blood cultures: early (< four hours of detection) and late (> four hours of detection). We focused on the early false-positives as they were far more significant
Author information
Authors and Affiliations
Contributions
Camille Petit: methodology, investigation, data curation, validation, formal analysis, visualization, writing—original draft preparation. Philippe Lavrard-Meyer: data curation, validation, visualization, writing—original draft preparation. Didier Raoult: conceptualization, methodology, investigation, validation writing—reviewing and editing. Grégory Dubourg: conceptualization, methodology, investigation, data curation, validation, writing—reviewing and editing.
Corresponding author
Ethics declarations
Ethics approval
This study was registered in the RGPD/AP-HM register under number 2021–98.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
D.R. is a scientific board member of Eurofins company, a founder of a microbial culture company (Culture Top), and was a consultant for Hitachi High-Technologies Corporation, Tokyo, Japan, from 2018. Funding sources had no role in the design and conduct of the study, the collection, management, analysis, and interpretation of the data, nor in the preparation, review, or approval of the manuscript.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Petit, C., Lavrard-Meyer, P., Raoult, D. et al. The absolute number of leukocytes per vial as a major cause of early false positive blood cultures: proof-of-concept and application. Eur J Clin Microbiol Infect Dis 41, 951–959 (2022). https://doi.org/10.1007/s10096-022-04454-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-022-04454-z