Abstract
Pneumocystis jirovecii pneumonia (PJP) is difficult to be diagnosed, so this study explored if PJP could be diagnosed by metagenomic next-generation sequencing (mNGS) and if mNGS could guide the therapy of PJP. mNGS was successfully diagnosed 13 out of 14 PJP recipients with 11 through peripheral blood samples, verified by PCR. Ten non-PJP recipients were enrolled as the control group. Blood tests revealed a high β-D-glucan (BDG) level in all recipients with PJP during the hospitalization. Four (28.6%) of 14 PJP patients were infected with cytomegalovirus simultaneously, while 8 (57.1%) suffered from a combined infection caused by Torque teno virus. Five (35.7%) of 14 cases died of PJP or the subsequent bacteremias/bacterial pneumonia with a longer interval between the onset and diagnosis of/the available therapy against PJP than survival cases. Univariate analysis of characteristics between PJP and non-PJP recipients revealed that BDG assays was higher at the admission in PJP group (P =0.011). This present study supports the value of mNGS detection of blood sample in diagnosing PJP, which could assist clinical decision for therapy against PJ and improve outcome of PJP. The study also highlights the sensitivity of BDG assays. Cytomegalovirus and Torque teno virus infections often occur at the same time of PJP, thus can be alerts of PJP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pneumocystis jirovecii pneumonia (PJP) is a serious and even critical opportunistic infection, which usually occurs in renal transplant recipients (RTRs) who are prone to respiratory infections, accompanied by multiple infections [1]. PJP cases have been reported frequently in solid organ transplant recipients, particularly in RTRs [2,3,4,5].
Precise and early diagnosis is important for the prognosis and survival of immunocompromised, critically ill RTRs. Nevertheless, as Pneumocystis jirovecii (PJ) cannot be reliably grown in vitro, PJP is diagnosed mainly through microscopic detection in respiratory specimens followed by staining methods, which generally shows low sensitivity and specificity [3, 6]. Detection of serum β-D-glucan (BDG) is sensitive but lack specificity [7]. Polymerase chain reaction (PCR)-based diagnostic assays have significantly improved the sensitivity and specificity, whereas its commercial kits have not yet been approved for clinical use in China [8].
Recently, metagenomic next-generation sequencing (mNGS), as an all-in-one diagnostic tool, has been successfully used for various infectious diseases, showing a higher detection rate of PJP compared to conventional methods [1, 9,10,11]. Of note, 4 cases of PJP were reported to be confirmed by mNGS in peripheral blood rather than respiratory samples [1, 12]. Although mNGS can diagnose PJP rapidly and noninvasively in immunocompromised patients, it was used to diagnose PJP only in 2 RTRs, awaiting further evaluation for its value [1, 2]. Herein we studied 14 cases of RTRs with PJP and 10 non-PJP RTRs predominantly identified by mNGS detection of blood samples rather than bronchoalveolar lavage fluid (BALF)/sputum to further explore its accuracy.
Materials and methods
Study design and study subjects
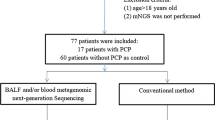
This was a single-center retrospective cohort study from the Third Xiangya Hospital, Central South University, China. All RTRs with a diagnosis of pneumonia admitted to our center from March 1, 2019, to October 31, 2020, were enrolled, divided into two groups: PJP or non-PJP. Clinical data of RTRs diagnosed with PJP or non-PJP mainly by mNGS and confirmed by PCR were analyzed. All patients included were at least 18 years old at the time of renal transplantation. They did not receive anti-PJ prophylaxis as a routine. No organs from executed prisoners were used. This retrospective non-interventional study involved 14 RTRs with PJP and 10 non-PJP RTRs, and these 14 RTRs did not receive any prophylaxis for PJ after transplantation. This study was approved by the Research Ethics Committee at the Third Xiangya Hospital of Central South University, consistent with principles in the Declaration of Helsinki (protocol no. IRB-2020-S636). The ethics committee waived the need for informed consent.
Routine immunosuppression
Induction immunosuppression consisted of basiliximab and/or antithymocyte globulin and methylprednisolone (0.5g given during the operative procedure, 200–250mg on postoperative day 1–3). Maintenance immunosuppression included mycophenolate mofetil, prednisone, and a calcineurin inhibitor, tacrolimus, or cyclosporin A.
Sample collection and processing
In our hospital, we have not performed urinary test antigens to discard S. pneumoniae or Legionella pneumophila infection yet. Routine cultures for bacteria and fungi were performed for all samples. A total volume of 8 ml blood samples was collected in BD vacutainer plasma preparation tubes for viral and microbial analysis using mNGS, or 3 ml BALF/sputum samples was provided for mNGS detection. Blood samples were stored at room temperature, and BALF/sputum samples were store with ice bags at 2–8 degrees Celsius. We only detected bacteria, fungi, and DNA viruses rather than RNA viruses by mNGS to save expenses and time, so we did not study the possibility of other RNA viral etiologies, such as influenza virus and respiratory syncytial virus.
DNA extraction, library construction, and sequencing
Viscous BALF or sputum samples were homogenized with normal saline. Blood samples were centrifuged at 1600 rpm, and BALF/sputum samples were centrifuged at 12,000 rpm for 10 min at 4°C to eliminate debris. Pyrolysis liquid GHL was added to release total DNA for BALF/sputum samples. Cell-free DNA from plasma and total DNA from BALF/sputum samples were extracted and purified using a nucleic acid extraction (DNA) or purification kit (Sansure Biotech, Magnetic beads method, S1005). Extracted DNA was measured by Qubit dsDNA HS Assay Kits (Thermo Fisher Scientific, Art. No. Q32854), and the total mass of DNA needs to be more than 1 ng as a quality control. The total DNA was further fragmented using dsDNA fragmentase. DNA libraries for sequencing were then constructed. The 5′ end of DNA fragment was phosphorylated, and a Da tail was added at the 3′ end, following adapters that were attached to the nucleic acid templates for subsequent sequencing and sample identification. Sequencing libraries were pooled with environmental control samples, which were used to monitor sample-to-sample contamination and were processed alongside the test samples in every batch. Pooled sample libraries up to 18 libraries per batch were multiplexed and sequenced on Illumina NextSeq500 sequencers using a 75-cycle single-end, single index sequencing kit.
Bioinformatic analysis
At least 20M reads were obtained for each respiratory sample, and up to 50M reads were obtained for blood sample. By removing low quality and adapter sequences using Trimmomatic, more than 99.95% of the original reads were used for analysis (so-called clean reads). More than 99% of the clean reads could be mapped to human genome (containing GRCh38 and Homo sapiens clone satellite sequences) using Burrows-Wheeler Alignment tool, so less than 1% of the clean reads was used for further analysis (so-called unmapped reads). A mixed single-end sequencing mode of SE75 or SE150 was used for the analysis. The full range of 75 or 150 bps was used for analysis. Subsequently, the remaining data were aligned to the bacterial, viral, fungal, and parasite databases using Burrows-Wheeler Alignment tool. The reference databases were downloaded from NCBI RefSeq (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq) and Genbank (ftp://ftp.ncbi.nlm.nih.gov/genomes/ genbank). The bacterial database contained 6600 different species, the fungal database contained 980 different species, the viral database contained 5000 different species, and parasite database contained 220 different species. The mapping standard criteria included the following: (1) continuous match numbers were more than 50bp. (2) Mismatch numbers were less than 3bp.
The mapped reads were aligned to the NCBI-nt database using Blastn for verification and filtration. We used the local Blastn by creating our own BLAST nucleotide database. It uses multi threads (threads=8) to run the blastn and it is faster than blastn online (threads=1). More information can be found in BLAST Manual (https://www.ncbi.nlm.nih.gov/books/ NBK279690/). We further calculated the coverage and abundance of species using Samtools. Some nonspecific reads would be filtered if they were mapped to different genus and different species within the standard mapping criteria. Only the reads mapped to unique species could be classified to the species level. According to the classification results, we obtained the information of genus and species for the tested samples.
Criteria for reporting a positive mNGS result
The database curated from books was used for pathogenic comments, which was after the positives were selected [13, 14]. The background database (containing species with detection rate greater than 50% in all whole blood samples) and the environmental control were used for filtering out the environmental contaminations. The criteria for reporting positive pathogens would be as follows:
-
1.
For fungi and viruses, pathogens with the mapping read number greater than or equal to 3 and the confidence level more than 99% will be selected.
-
2.
For bacteria (Mycobacterium tuberculosis excluded) and parasites, the pathogens will be reported if the mapping read number was in the top 5 of bacteria or parasitic list and not in the background library [15].
-
3.
Mycobacterium tuberculosis would be considered positive if at least 1 read was mapped to either the species or genus level, due to the difficulty of DNA extraction and low possibility of contamination [16].
PCR validation
Sterile centrifuge tubes were used to collect blood, sputum, or alveolar lavage fluid from patients. The sample volume after liquefaction should not be less than 1mL. The sample to be tested could be placed at 2~8°C for less than 72 h. Long-term storage should be placed under the condition of ≤−20°C to avoid repeated freezing and thawing. The specimens were transported in a curling pot with ice or a foam box with ice sealed for transportation.
Two hundred microliters of the sample to be tested was put into a 1.5-mL centrifuge tube, to perform nucleic acid extraction using the nucleic acid extraction or purification reagent (S1006, Sansure Biotech) according to its instruction.
The PCR validation was carried out using the PJ nucleic acid detection kit (PCR-fluorescent probe method, Sansure Biotech). The fluorescence detection used the ROX channel to detect PJ and the CY5 channel to detect the internal standard.
If the ROX channel detected a typical S-type amplification curve, and Ct ≤ 40, the PJ was positive. If Ct > 40 or no Ct, it was negative. If the internal standard did not detect Ct or Ct > 40 in the CY5 channel, which meant that the concentration of the test sample was too low or there were interfering substances that inhibited the reaction, the experiment was repeated again.
The methods for other tests
Immunoturbidimetric methods were used for the C-reactive protein and BDG quantification, and electrical impedance methods were used for WBC count. Procalcitonin was detected by chemiluminescence technique.
Statistical analysis
All analyses were performed using the statistical package SPSS for Windows, version 22.0 (IBM Corporation, Armonk, NY, USA). Results were listed as mean (±SD) for continuous variables. Univariate analysis was applied to inspect the distinction of characteristics between PJP and non-PJP groups. Continuous variables were compared using Student’s unpaired t-test. Categorical variables were compared with Pearson’s χ2 test or Fisher’s exact test. Statistical tests were two-tailed, and statistical significance was defined as P<0.05.
Results
There were 14 RTRs diagnosed with PJP in our center from March 1, 2019, to October 31, 2020. The baseline characteristics of the 14 PJP recipients enrolled were presented in Table 1. Time interval between renal transplantation and PJP was less than 3 years. The median age was 37.6 (range, 19–54) years. Ten participants were males. Seven recipients had renal insufficiency, and 2 suffered from diabetes at the admission. All 14 recipients received chest high-resolution computed tomography scans at the admission, suggesting reticular and/or ground-glass opacities pulmonary infiltrates without pleural effusion, consistent with characteristics of interstitial pneumonia. During hospitalization, 7 recipients progressed to severe acute respiratory distress syndrome (ARDS), and acute renal failure occurred in 1. All patients were initially prescribed 3 maintained immunosuppressants (tacrolimus/cyclosporin A + mycophenolate mofetil + glucocorticosteroids), and additional agents of basiliximab and/or antithymocyte globulin were administrated as an induction therapy in 10 recipients.
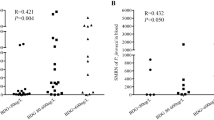
Five of 14 (35.7%) cases died: one each died of PJP, the subsequent bacterial pneumonia caused by A. baumannii, and the subsequent bacteremia due to A. baumannii, and 2 died of the subsequent bacteremia owing to K. pneumoniae. The mean time interval between the onset and the diagnosis of PJP in these five died patients was 24 (ranged from 9 to 40) days, with 16.4 (ranged from 7 to 35) days of the mean interval between the onset of PJP and the available therapy against PJP. The corresponding intervals were only 9 (ranged from 6 to 18) and 4.9 (ranged from 2 to 11) days in 9 survival recipients. Treated with trimethoprim-sulfamethoxazole and a certain time period of caspofungin for 15–49 days, these 9 RTRs were cured and discharged. Laboratory tests and mNGS results obtained from the 14 RTRs with PJP were listed in Table 2. We conducted mNGS in 21 samples including 16 peripheral blood and 5 respiratory samples collected from enrolled recipients. The respiratory samples consisted of 3 BALF and 2 sputum samples. All the cases were receiving antibiotics at the time of sample collection for mNGS, with 10 receiving other antifungal agents at the same time and 1 receiving agents actively against PJ as long as 11 days. Two patients had leukopenia (white blood cell count <4000/μL) and 6 had severe lymphopenias (absolute lymphocyte count < 500/μL) at baseline. C-reactive protein was more than 60 mg/L in 57.1% of patients (8/14), while procalcitonin was relatively low in all patients (ranged from 0.019 to 1.567 ng/mL) at baseline. All 14 patients underwent 58 times of BDG assays with 50 (86.2%) times being positive. Eight negative results appeared either early in the onset of the disease (n=1) or near recovery (n=7). Among 50 times of BDG assays, 29 times were more than 400 pg/mL.
Thirteen (92.9%) of 14 RTRs were diagnosed with PJP by mNGS, verified by PJ PCR, consistent with clinical manifestations (fever, cough, dyspnea, and mostly normal pulmonary auscultation even with significant hypoxemia) and radiological findings (diffuse ground-glass opacity with patchy distribution, cysts, grid-like, or pavement pebble-like performance) of interstitial pneumonia. Eleven of 14 patients provided peripheral blood samples. Of them, 2 simultaneously provided respiratory samples. The other 3 cases used BALF or sputum samples. Notably, the PJ sequencing reads fluctuated as the infection state changed in Patient 5 who underwent five times of mNGS detection of blood sample, and eventually no PJ was detected before his discharge. Microscopic examination of stained sputum found PJ in 1 case. Ct values of PJ PCR of these 13 recipients with mNGS positive ranged from 25.14 to 38.42. The Ct value of the remaining 1 PJP recipient was more than 40.
Mixed infections were observed in 10 PJP recipients. mNGS identified cytomegalovirus in 4 cases (3 of them cytomegalovirus replication confirmed by standard PCR); Torque teno virus in 8 cases; Candida parapsilosis in 2 cases; and Pseudomonas putida, Haemophilus parainfluenzae, Mycoplasma orale, BK virus, Human polyomavirus 8, Human polyomavirus 1, and Human coronavirus 229E in 1 case each. Furthermore, A. baumannii was discovered in blood and BALF cultures in Patient 2 and in sputum culture in Patient 12 within 2 days before their death. E. faecium in two urine cultures was found within 8 days while A. baumannii in BALF culture and K. pneumoniae in both BALF and blood cultures within 2 days before death of Patient 3. K. pneumoniae in both BALF and blood cultures was found within 2 days before death of Patient 14.
mNGS identified PJP in 7 RTRs (Patients 4–10) with culture-negative sepsis. They had compatible clinical syndromes and responded well to the therapy against PJ. Patient 4 underwent renal transplantation 5 months and developed fever and dyspnea 3 days before the admission with CT confirmed ground-glass opacity and grid-like performance and with pulmonary dry rales. She was treated with trimethoprim-sulfamethoxazole (4320–7680 mg/day) for 18 days and caspofungin (50–70 mg daily) for 3 days and was cured and discharged. Patient 5 underwent renal transplantation 2 months, and fever occurred 3 days before the admission with CT confirmed ground-glass opacity and grid-like performance and with normal pulmonary auscultation. He was treated with trimethoprim-sulfamethoxazole (5760–7680 mg/day) and caspofungin (70 mg loading dose followed by 50 mg/day maintenance dose) for 23 days and was cured. Patient 6 underwent renal transplantation 4 months, and fever and cough developed 4 days before the admission with CT showing interstitial pneumonia and with normal pulmonary auscultation. She was treated with trimethoprim-sulfamethoxazole (1920 mg/day) for 30 days and caspofungin (70 mg loading dose followed by 50 mg/day maintenance dose) for 25 days and was cured and discharged. Patient 7 underwent renal transplantation 3 months, and fever developed 1 day before the admission with normal pulmonary auscultation and with CT showing diffuse ground-glass opacity with patchy distribution. He was treated with trimethoprim-sulfamethoxazole (1920–2880 mg/day) for 24 days and caspofungin (70 mg loading dose followed by 50 mg/day maintenance dose) for 4 days and was cured and discharged. Patient 8 underwent renal transplantation 3 months, and fever, cough, and sputum developed 7 days before the admission with pulmonary moist rales and with CT showing diffuse ground-glass opacity with patchy distribution, scattered small nodular shadow in the right lung, and cysts in the left lower lung. Trimethoprim-sulfamethoxazole (1920–3640 mg/day) was prescribed to him for 42 days. Patient 9 underwent renal transplantation 7 months and presented fever 4 days before the admission with normal pulmonary auscultation and with CT showing interstitial pneumonia. He was treated with trimethoprim-sulfamethoxazole (2880–7680 mg/day) for 25 days and caspofungin (70 mg loading dose followed by 50 mg/day maintenance dose) for 13 days and was cured. Patient 10 underwent renal transplantation 2 years and fever and cough occurred 3 days before the hospitalization with normal pulmonary auscultation and with CT showing diffuse ground-glass opacity, high-density patches, and solid nodules in the right lung. He was treated with trimethoprim-sulfamethoxazole (2880–5760 mg/day) for 19 days and caspofungin (70 mg loading dose followed by 50 mg/day maintenance dose) for 17 days and was cured.
Ten non-PJP recipients with mNGS negative were enrolled as the control group. The Ct values of all these 10 non-PJP recipient were more than 40. None of these patients had the clinical manifestations and radiological findings of interstitial pneumonia. The baseline characteristics of them were presented in Table 3. All of them were diagnosed by mNGS, 9 of whom were detected through peripheral blood with 1 through the combination of blood and sputum, and the remaining 1 recipient was detected through sputum. The median age was 43.1 (range, 19–64) years. Eight participants were males. Five recipients had renal insufficiency, and 2 suffered from diabetes at the admission. All patients were initially prescribed 3 maintained immunosuppressants (tacrolimus + mycophenolate mofetil + glucocorticosteroids), and additional agents of basiliximab and/or antithymocyte globulin were administrated as an induction therapy in 5 recipients. During hospitalization, 2 recipients progressed to ARDS, and one died of pulmonary embolism.
A number of variables, including age, gender, severe ARDS, BDG assays at the admission, white blood cell/lymphocyte, C-reactive protein/procalcitonin at the admission, and underlying disease such as renal insufficiency and diabetes, were compared between groups of PJP (n=14) and non-PJP (n=10) by univariate analysis. Only 1 non-PJP recipient who suffered from the infection caused by Candida parapsilosis had a high BDG level at the admission. As shown in Table 4, only the outcome of BDG assays, including the absolute value (P=0.011) and positive rate (P<0.001), was associated with the occurrence of PJP by univariate analysis, suggesting that BDG value at the admission in PJP patients was higher than that in non-PJP patients. Mortality rates between two groups were also compared by univariate analysis, but no significant difference was found, although the rate was higher in PJP recipients than that in non-PJP cases (35.7% vs. 10.0%; P=0.151).
Discussion
PJP is a severe fungal infection and has a high prevalence in immunocompromised individuals, thus posing a huge threat to these patients. Delayed diagnosis of all invasive fungal infection associates with higher mortality [17]. Diagnosis of PJP remains challenging due to its nonspecific clinical manifestations as well as low sensitivity and specificity of current conventional diagnostic methods [2]. Up till now, the definitive diagnosis of PJP relies on microscopic examination of stained sputum smears, lung tissues, or BALF samples. Wright-Giemsa and methenamine silver-stained smear are the most commonly applied methods in past decades. Fluorescence staining with monoclonal antibodies could increase the detection sensitivity to 95% [18]. PCR and serologic biomarkers such as serum BDG or S-adenosyl methionine level were also used to increase the PJP diagnostic rate in recent years [1, 19]. However, the efficiency of serum BDG test for diagnosing PJP is controversial. Some studies claimed that it lacked sensitivity and specificity [1, 2], while in 3 meta-analyses, it was demonstrated to have a sensitivity of 91 to 96%, which is similar to the 92.9% (13/14) sensitivity of BDG in our present study at the admission [20,21,22]. We also found that the value of BDG assays at the admission was higher and was more likely positive in PJP patients than that in non-PJP patients by univariate analysis, suggesting the high sensitivity of BDG assays for PJP once again.
Compared to PCR and other conventional diagnostic methods, mNGS is a comprehensive approach, allowing sequence-based identification of all potential pathogenic microbes in clinical samples with minimized turn-around time of less than 48 h [23, 24]. In the present study, the mNGS analysis report could be obtained within 24–48 h starting from the samples received in the laboratory. To date, there have been several reports on successful use of mNGS to diagnose pulmonary infections involving PJ, with two of them reporting that mNGS is more sensitive than conventional methods (non-PCR based) [1, 2, 9,10,11,12]. The sensitivity (90%) and specificity (100%) of mNGS are consistent with the PCR test in our results. The mNGS is much slower and more expensive than a PCR test. However, the PCR test requires preknowledge or suspicion of a specific infection such as PCP. mNGS covers a broad spectrum of infections or pathogens, and it helps to identify specific pathogens for most unexpected cases. Under life-threatening situations, mNGS could save time and life by identifying unexpected infections.
It is surprising that in Patient 1, sputum methenamine silver-stained smear was performed, and PJ was found 6 days after the blood mNGS which did not reveal PJ infection. The reason for this discrepancy may be an early stage of the disease when a blood mNGS was performed and PJ has not entered blood yet. In the present study, in PJP Patient 11 with a positive BALF culture, Pseudomonas putida grown was detected by BALF mNGS on the same day, and in non-PJP Patient 8, the same pathogen Pseudomonas aeruginosa was detected on blood culture and mNGS with blood culture 2 days prior to blood mNGS. It is not surprising that in PJP Patients 2, 3, and 14 with a positive blood culture, the organism grown was not detected by blood mNGS since the interval between blood culture and blood mNGS was at least 3 days.
What is more, mNGS is able to detect organisms in the setting of effective treatment, which was proved by our present study where Patient 2 and 5 had received 11 and 13 days of active therapy against PJ prior to the collection of BALF sample and the fourth blood sample for mNGS testing, respectively [12].
PJP has been confirmed in 4 cases by mNGS in peripheral blood rather than respiratory samples in only 2 previous studies [1, 12]. Although PJ almost exclusively inhabits the human lungs, pieces of PJ could penetrate through respiratory infectious site and circulate into peripheral blood, especially in immune-suppressed patients; thus, blood PJ sequence reads can diagnose PJP [1, 6]. There were only two other studies involving 2 RTRs which utilized mNGS to detect BALF samples and find PJ [1, 2]. Herein we found that mNGS successfully detected PJ in 10 out of 11 PJP who provided blood samples, verified by PJ PCR, highlighting the value of mNGS of blood in diagnosing PJP. In addition, obtaining high-quality blood samples is easier than BALF samples. In our present study, although Patients 6, 9, and 13 had very low PJ reads of 9, 7, and 6 in blood mNGS and Ct values >35 in PCR. However, PJ in blood can only originate from the lungs. Furthermore, if the ROX channel detected a typical S-type amplification curve, and Ct ≤ 40, PJ was positive. This is the reason why we still think Patients 6, 9, and 13 had a PJP. Nevertheless, with the limit of sample size, larger clinical trials investigation on evaluating blood mNGS for PJP diagnosis among RTRs is necessary.
The mean time interval between the onset and diagnosis of/the available therapy against PJP in died group was substantially longer than survival group. The difference of the time interval between the onset and diagnosis of PJP or between the onset of PJP and beginning of antibiotic treatment so different between died and survivor patients with PJP just reflected the low diagnostic rate of PJP because commercial PCR kits for PJ have not yet been approved for clinical use in China as well as the randomness of the treatment for PJP in China due to no guideline on how to prevent PJP among transplant recipients in China. Our present report highlights the need to include PJP prophylaxis in the relevant guideline in China.
Furthermore, in our present study, mNGS rapidly identified PJP diagnosis and effectively guide the therapy against PJ in 7 recipients with culture-negative pneumonia. In addition, mNGS revealed the frequent combined infection of PJ with cytomegalovirus or Torque teno virus. Torque teno virus belongs to Anelloviridae family and lacks pathogenic potential and relevant clinical manifestation so far [12, 25]. It is considered to mirror T cell function as well as humoral and innate immunity and is, therefore, a surrogate marker of heavy immunodeficiency [26,27,28,29]. The prevalence of Torque teno virus in solid organ recipients is up to 99% [30].
In conclusion, the incidence of PJP was high, with 14 cases occurring in RTRs over less than 2-year period in our center. This study supports the value of mNGS in guiding early treatment for PJP and helping exclusion of infection with minimized turn-around time and improved sensitivity and specificity in diagnosing PJP, even in detecting organisms in the setting of effective treatment. mNGS can improve the outcome of RTRs with PJP since the longer interval between the onset and diagnosis of/the available therapy against PJP in died group. Moreover, mNGS showed great diagnostic performance in multiple organisms. mNGS of blood samples could be an alternative choice particularly for patients with pneumonia of unknown etiology in whom bronchoscopy or biopsy for tissue diagnosis is not feasible. This study also highlights the sensitivity of BDG assays for PJP. PJP often co-occurs with cytomegalovirus or Torque teno virus infections, making them alerts of PJP.
Data availability
The data that support the findings of this study are available within the present paper.
References
Zhang Y, Ai JW, Cui P et al (2019) A cluster of cases of Pneumocystis pneumonia identified by shotgun metagenomics approach. J Inf Secur 78:158–169
Chen J, He T, Li X et al (2020) Metagenomic next-generation sequencing in diagnosis of a case of pneumocystis jirovecii pneumonia in a kidney transplant recipient and literature review. Infect Drug Resist 13:2829–2836
Le Gal S, Toubas D, Totet A et al (2020) Pneumocystis infection outbreaks in organ transplantation units in France: a nation-wide survey. Clin Infect Dis 70:2216–2220
de Boer MG, de Fijter JW, Kroon FP (2011) Outbreaks and clustering of Pneumocystis pneumonia in kidney transplant recipients: a systematic review. Med Mycol 49:673–680
Yiannakis EP, Boswell TC (2016) Systematic review of outbreaks of Pneumocystis jirovecii pneumonia: evidence that P. jirovecii is a transmissible organism and the implications for healthcare infection control. J Hosp Infect 93:1–8
Ma L, Cisse OH, Kovacs JA (2018) A molecular window into the biology and epidemiology of Pneumocystis spp. Clin Microbiol Rev 31:e00009–e00018
Del Corpo O, Butler-Laporte G, Sheppard DC et al (2020) Diagnostic accuracy of serum (1-3)-β- D-glucan for Pneumocystis jirovecii pneumonia: a systematic review and meta-analysis. Clin Microbiol Infect 26:1137–1143
Summah H, Zhu YG, Falagas ME et al (2013) Use of real-time polymerase chain reaction for the diagnosis of Pneumocystis pneumonia in immunocompromised patients: a meta-analysis. Chin Med J 126:1965–1973
Wang J, Han Y, Feng J (2019) Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med 19:252
Irinyi L, Hu Y, Hoang MTV et al (2020) Long-read sequencing based clinical metagenomics for the detection and confirmation of Pneumocystis jirovecii directly from clinical specimens: a paradigm shift in mycological diagnostics. Med Mycol 58(5):650–660
Li Y, Sun B, Tang X et al (2020) Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients. Eur J Clin Microbiol Infect Dis 39:369–374
Camargo JF, Ahmed AA, Lindner MS et al (2019) Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000Res 8:1194
Kasper D, Fauci A (2016) Harrison’s infectious diseases, 3/E. McGraw-Hill Medical
Jorgensen JH (2015) Manual of Clinical Microbiology, 11th edn. ASM Press
Langelier C, Fung M, Caldera S et al (2020) Detection of pneumonia pathogens from plasma cell-free DNA. Am J Respir Crit Care Med 201:491–495
Miao Q, Ma Y, Wang Q et al (2018) Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis 67(suppl_2):S231–S240
Morrell M, Fraser VJ, Kollef MH (2005) Delaying the empiric treatment of candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother 49:3640–3645
Limper AH, Adenis A, Le T et al (2017) Fungal infections in HIV/AIDS. Lancet Infect Dis 17:e334–e343
de Boer MG, Gelinck LB, van Zelst BD et al (2011) beta-D-glucan and S-adenosylmethionine serum levels for the diagnosis of Pneumocystis pneumonia in HIV-negative patients: a prospective study. J Infect Jan 62:93–100
Onishi A, Sugiyama D, Kogata Y et al (2012) Diagnostic accuracy of serum 1,3-beta-D-glucan for Pneumocystis jiroveci pneumonia, invasive candidiasis, and invasive aspergillosis: systematic review and meta-analysis. J Clin Microbiol 50:7–15
Karageorgopoulos DE, Qu JM, Korbila IP, Zhu YG et al (2013) Accuracy of beta-D-glucan for the diagnosis of Pneumocystis jirovecii pneumonia: a meta-analysis. Clin Microbiol Infect 19:39–49
Li WJ, Guo YL, Liu TJ et al (2015) Diagnosis of Pneumocystis pneumonia using serum (1-3)-beta-D-glucan: a bivariate meta-analysis and systematic review. J Thorac Dis 7:2214–2225
Chiu CY, Miller SA (2019) Clinical metagenomics. Nat Rev Genet 20:341–355
Somasekar S, Lee D, Rule J et al (2017) Viral surveillance in serum samples from patients with acute liver failure by metagenomic next-generation sequencing. Clin Infect Dis 65:1477–1485
Lewandowska DW, Schreiber PW, Schuurmans MM et al (2017) Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection. PLoS One 12:e0177340
Schiemann M, Puchhammer-Stöckl E, Eskandary F et al (2017) Torque teno virus load-inverse association with antibody-mediated rejection after kidney transplantation. Transplantation. 101:360–367
Martín-López M, Albert E, Fernández-Ruiz M et al (2020) Torque teno virus viremia in patients with chronic arthritis: influence of biologic therapies. Semin Arthritis Rheum 50:166–171
Giacconi R, Maggi F, Macera L et al (2018) Torquetenovirus (TTV) load is associated with mortality in Italian elderly subjects. Exp Gerontol 112:103–111
Freer G, Maggi F, Pifferi M et al (2018) The virome and its major component, anellovirus, a convoluted system molding human immune defenses and possibly affecting the development of asthma and respiratory diseases in childhood. Front Microbiol 9:686
Gorzer I, Jaksch P, Kundi M et al (2015) Pre-transplant plasma torque teno virus load and increase dynamics after lung transplantation. PLoS One 10:e0122975
Funding
This work was support by the Science and Technology Department of Hunan Province, China (grant 2020JJ4851).
Author information
Authors and Affiliations
Contributions
ZF and WQQ participated in research design and drafted the manuscript. CJ, HH, and WQQ participated in patient enrollment. ZF and CJ performed the statistics. LRJ was responsible for the implementation of overall bioinformatics and mNGS results. ZW did the experimental work of mNGS. ZM was responsible for the workflow and interpretation of mNGS results. DXL performed the bioinformatics analysis of mNGS results. DLZ was responsible for the invention and implementation of nucleic acid extraction in both mNGS and PCR. All authors participated in the writing and revision of the manuscript and accepted the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, F., Chen, J., Huang, H. et al. Application of metagenomic next-generation sequencing in the diagnosis and treatment guidance of Pneumocystis jirovecii pneumonia in renal transplant recipients. Eur J Clin Microbiol Infect Dis 40, 1933–1942 (2021). https://doi.org/10.1007/s10096-021-04254-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04254-x