Abstract
We evaluated a novel transcription-reverse transcription concerted reaction (TRC) assay that can detect influenza A and B within 15 min using nasopharyngeal swab and gargle samples obtained from patients with influenza-like illness, between January and March 2018 and between January and March 2019. Based on the combined RT-PCR and sequencing results, in the nasal swabs, the sensitivity and specificity of TRC for detecting influenza were calculated as 1.000 and 1.000, respectively. In the gargle samples, the sensitivity and specificity of TRC were 0.946 and 1.000, respectively. The TRC assay showed comparable performance to RT-PCR in the detection of influenza viruses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transcription-reverse transcription concerted reaction (TRC) method is a combination of direct and rapid isothermal RNA amplification and real-time identification using an intercalation-activating fluorescence (INAF) probe [1, 2]. In Europe, Japan, and Vietnam, TRC ready-to use regents have been used for the diagnosis of tuberculosis [3], nontuberculous mycobacterial infections, Chlamydia infection, gonorrhea, and mycoplasma pneumonia. Recently, a novel TRC assay that can detect influenza A and B within 15 min was developed [4, 5]. In this study, we evaluated the efficacy of the novel rapid TRC assay for detecting influenza viruses in nasopharyngeal swab and gargle samples obtained from patients with influenza-like illness.
Materials and methods
Ethics
This study was approved by the ethics committee of Nagasaki University Hospital (approval numbers: 17121822 and 18111919) and was registered at the UMIN Clinical Trials Registry (reference numbers: UMIN000032395 and UMIN000034545). Written informed consent for participation in and publication of this study was obtained from all participants before sample collection.
Study design
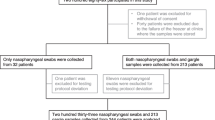
A prospective observational study was conducted in period 1, between January and March 2018, and period 2, between January and March 2019. Patients who visited or were hospitalized at the Department of Respiratory Medicine, Japanese Red Cross Nagasaki Genbaku Hospital, with influenza-like illness (ILI) [6] were included in this study. Patients were excluded if they were administered anti-influenza agents within 1 month prior to the day on which they were sampled. Nasopharyngeal swabs were collected from patients in both periods using two swabs: one was used for antigen testing using silver amplification immunochromatography (FUJI DRI-CHEM IMMUNO AG Cartridge FluAB, Fujifilm, Kanagawa, Japan) [7] at Nagasaki Genbaku Hospital and the other was stored at −20 °C until further analysis. Gargle samples were collected from patients in period 2. For gargle sampling, the patients gargled their throat with 20 mL distilled water for 10 s. The physicians determined the clinical diagnosis with history taking, physical findings, and results of the influenza antigen test. All information, such as clinical report forms and results of the TRC and RT-PCR, was summarized and analyzed at Nagasaki University Hospital.
Sample analysis
We performed a rapid TRC assay based on the protocol described in Japan-patent (JP,2017-195871, A) at Nagasaki University Hospital. In summary, the procedure was performed as follows. A nasopharyngeal swab or gargle swab was mixed in 1-mL extraction buffer containing surfactant and incubated at 52 °C for 1 min. Thirty microliters of the sample was mixed with dry reagent containing enzymes, substrates, primers, and INAF probes. The mixture was incubated at 46 °C, and the fluorescence was monitored. RT-PCR and sequencing were performed as a gold standard at Tosoh Corporation based on the Influenza Diagnosis Manual [8,9,10]. Total RNA was isolated from 140 μL of TRC extraction buffer mixed with a nasopharyngeal swab or 140 μL of a gargle specimen using the Qiagen RNeasy kit. RT-PCR was performed using the One Step PrimeScript™ RT-PCR Kit (TAKARA BIO, Shiga, Japan). TRC assay and RT-PCR were performed independently, and the results of them were combined at Nagasaki University Hospital after all analysis was completed. If the results of the TRC and RT-PCR were different, the samples were analyzed by sequencing.
Statistical analysis
All statistical analyses were performed with EZR version 1.41 (Saitama Medical Center, Jichi Medical University, Saitama, Japan) [11], which is a graphical user interface for R (the R Foundation for Statistical Computing, Vienna, Austria; version 3.6.3). Fisher’s exact test was used to compare categorical variables, and the statistical significance level was set at <0.05. The sensitivity (Se), specificity (Sp), positive predictive value (PPV), and negative predictive value (NPV) of the TRC against the combined results of the RT-PCR and sequencing were calculated with 95% confidence intervals (95% CI).
Results
Patient characteristics
During the study period, a total of 188 patients were evaluated, comprising 92 patients in period 1 and 96 patients in period 2. Patient characteristics are shown in Table 1. Of the patients, 95 (50.5%) visited a hospital within 24 h of the onset of symptoms. The percentage of patients with fatigue was significantly lower in period 1 (37.0%) than in period 2 (71.9%, P = <0.001). The influenza antigen test using silver amplification immunochromatography detected influenza A and B in 38 (20.2%) and 39 (20.7%) patients, respectively. The percentage of influenza A was significantly lower, while that of influenza B was significantly higher, in period 1 than in period 2 (Table 1).
Comparison of TRC and RT-PCR results
The results of the RT-PCR and TRC are shown in Table 2. In the nasal swabs, influenza A and B were detected using RT-PCR in 36 (19.1%) and 39 (20.7%) patients, respectively, and were detected using TRC in 38 (20.2%) and 40 (21.3%) patients, respectively (Table 2). Of all patients, three tested negative for influenza with RT-PCR, but positive with TRC. Influenza A was detected in two of these three patients, and influenza B was detected in one by sequencing. Based on the combined RT-PCR and sequencing results, the Se, Sp, PPV, and NPV of the TRC were 1.000, 0.973, 0.962, and 1.000, respectively (Table 3).
In the gargle samples, influenza A was detected by RT-PCR and TRC in 37 (38.5%) and 35 (36.5%) patients, respectively (Table 2). Of all patients in period 2, two tested positive for influenza A with RT-PCR, but negative with TRC. Influenza A was detected in these two patients by sequence analysis. Based on the combined RT-PCR and sequence analysis results, the Se, Sp, PPV, and NPV of the TRC in the gargle samples were 0.946, 1.000, 1.000, and 0.967, respectively (Table 3).
Comparison of results between nasopharyngeal swabs and gargle samples
In the RT-PCR testing, five patients tested negative for influenza A in their nasopharyngeal swabs, but tested positive for influenza A in their gargle samples (Fig. 1). The Se, Sp, PPV, and NPV of the RT-PCR in the gargle samples were 1.000, 0.922, 0865, and 1.000, respectively. The Se, Sp, PPV, and NPV of the TRC in the gargle samples were 0.912, 0.935, 0.886, and 0.951, respectively.
Discussion
The novel rapid TRC assay showed great sensitivity and specificity in nasopharyngeal swabbing in both periods 1 and 2. Based on the results of the antigen test, period 1 was considered to be the influenza B epidemic season and period 2 was considered the influenza A epidemic season (Table 1). The sensitivity and specificity of the rapid TRC assay were 1.000 and 1.000, respectively, based on the combined RT-PCR and sequencing results for both periods. There are several rapid RT-PCR assays for detection of influenza, such as the ID Now influenza A and B 2 assay (ID Now), Cobas influenza A/B nucleic acid test (Liat), and Xpert Xpress Flu assay (Xpert). The previous studies reported that the sensitivity and specificity of these methods for detecting influenza A/B were 0.932 to 1.000/0.917 to 1000 and 0.977 to 1.000/0.976 to 0.998, respectively [12,13,14]. These results indicate that the performance of the rapid TRC assay is comparable to that of rapid RT-PCR assays.
In the present study, the rapid TRC assay also showed great sensitivity and specificity for gargle sampling. The rapid TRC assay and RT-PCR detected influenza in more patients from gargle samples than from nasopharyngeal swabs. The sensitivity and specificity of the Rapid TRC assay were 0.946 and 1.000, respectively. The previous studies reported that the sensitivity of Xpert and Liat for gargle sampling was 0.917 and 1.000, respectively, in comparison with in-house RT-PCR [15, 16]. Although there are no data on the specificity of rapid RT-PCR assays, the rapid TRC assay seems to be comparable with rapid RT-PCR assays for gargle samples. In the diagnosis of influenza, nasopharyngeal swabbing is the major sampling type, but these samples are difficult to obtain and the procedure is uncomfortable for patients [17]. In addition, due to the novel coronavirus (SARS-CoV-2) pandemic, healthcare workers must wear personal protective equipment when they obtain nasopharyngeal swabs. Therefore, it is vital to develop a diagnostic method, such as the rapid TRC assay, using gargle samples, that is easy to perform, non-invasive, material saving, and safe for healthcare workers [18].
There are some limitations in this study. First, the samples were obtained from one community hospital in Nagasaki, which might have limited the generalizability of the findings. Second, we used an equipment of the novel TRC assay under development. Accordingly, we are conducting a multicenter study for the rapid TRC assay using production version. Third, the rapid TRC assay was not compared with other rapid RT-PCR assays. Since these assays have not yet been approved by the Japanese Pharmaceuticals and Medical Devices Agency, we will conduct a comparative study in the future.
In conclusion, the novel rapid TRC assay showed comparable performance to RT-PCR in the detection of influenza viruses. In addition, because it could detect influenza viruses using gargle samples, the rapid TRC assay could contribute to the diagnosis of influenza during the SARS-CoV-2 pandemic.
Availability of data and materials
Raw data were generated at Nagasaki University Hospital. Derived data supporting the findings of this study are available from the corresponding author on request.
References
Ishiguro T, Saitoh J, Horie R, Hayashi T, Ishizuka T, Tsuchiya S et al (2003) Intercalation activating fluorescence DNA probe and its application to homogeneous quantification of a target sequence by isothermal sequence amplification in a closed vessel. Anal Biochem 314:77–86. https://doi.org/10.1016/S0003-2697(02)00618-8 [Internet]. [cited 2020 1]; https://www.sciencedirect.com/science/article/pii/S0003269702006188?via%3Dihub
Ishiguro T, Saitoh J, Yawata H, Otsuka M, Inoue T, Sugiura Y (1996) Fluorescence detection of specific sequence of nucleic acids by oxazole yellow-linked oligonucleotides. Homogeneous quantitative monitoring of in vitro transcription. Nucleic Acids Res 24:4992–4997. https://doi.org/10.1093/nar/24.24.4992 [Internet]. [cited 2020 1]; http://www.ncbi.nlm.nih.gov/pubmed/9016671
Mazzarelli A, Cannas A, Venditti C, D’Arezzo S, De Giuli C, Truffa S et al (2019) Clinical evaluation of TRCReady M.TB for rapid automated detection of M. tuberculosis complex in respiratory samples. Int J Tuberc Lung Dis 23:260–264. https://doi.org/10.5588/ijtld.18.0355 [Internet]. [cited 2020 1]; https://pubmed.ncbi.nlm.nih.gov/30808461/?from_single_result=Clinical+evaluation+of+TRC+Ready+for+rapid+automated+detection+of+complex+in+respiratory+samples&expanded_search_query=Clinical+evaluation+of+TRC+Ready+for+rapid+automated+detection+of+complex+i
Makino Y, Miki D, Noguchi Y, Sato H (2016) Primer for amplifying influenza virus RNA, and influenza virus detection method using the same. [Internet]. [cited 2020 1]; https://worldwide.espacenet.com/patent/search/family/056425709/publication/JP2016131498A?q=JP6565193B
Hatashita R, Makino Y, Miki D, Noguchi A, Sato H Reagents for extraction and amplification of nucleic acid from virus. [Internet]. [cited 2020 15]; https://www.j-platpat.inpit.go.jp/c1800/PU/JP-2017-195871/D3BADD8449A11E61DF2F46A85FCA8CF8E5102F972C4E673D83731053DB2C6584/11/en
World Health Organization Global Epidemiological Surveillance Standards for Influenza [Internet]. [cited 2020 10]. https://www.who.int/influenza/resources/documents/WHO_Epidemiological_Influenza_Surveillance_Standards_2014.pdf?ua=1
Mitamura K, Shimizu H, Yamazaki M, Ichikawa M, Nagai K, Katada J et al (2013) Clinical evaluation of highly sensitive silver amplification immunochromatography systems for rapid diagnosis of influenza. J Virol Methods 194:123–128. https://doi.org/10.1016/j.jviromet.2013.08.018 [Internet]. [cited 2019 2]; https://linkinghub.elsevier.com/retrieve/pii/S0166093413003509
National Institute of Infectious Diseases (2014) Manual for virus detection-influenza, 3rd edn. [Internet]. [cited 2020 15]; https://www.niid.go.jp/niid/images/lab-manual/Influenza2014.pdf
Furuse Y, Tamaki R, Okamoto M, Saito-Obata M, Suzuki A, Saito M et al (2019) Association between preceding viral respiratory infection and subsequent respiratory illnesses among children: a prospective cohort study in the Philippines. J Infect Dis 219:197–205. https://doi.org/10.1093/infdis/jiy515 [Internet]. [cited 2020 22]; http://www.ncbi.nlm.nih.gov/pubmed/30189092
Kyaw Win SM, Saito R, Win NC, Lasham DJ, Kyaw Y, Lin N et al (2020) Epidemic of influenza A(H1N1)pdm09 analyzed by full genome sequences and the first case of oseltamivir-resistant strain in Myanmar 2017. PLoS One 15:e0229601. https://doi.org/10.1371/journal.pone.0229601 [Internet]. [cited 2020 15]; https://dx.plos.org/10.1371/journal.pone.0229601
Kanda Y (2013) Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 48:452–458. https://doi.org/10.1038/bmt.2012.244 [Internet]. [cited 2020 16]; http://www.ncbi.nlm.nih.gov/pubmed/23208313
Kanwar N, Michael J, Doran K, Montgomery E, Selvarangan R (2020) Comparison of the ID now Influenza A & B 2, Cobas Influenza A/B, and Xpert Xpress flu point-of-care nucleic acid amplification tests for Influenza A/B virus detection in children. J Clin Microbiol 58. https://doi.org/10.1128/JCM.01611-19 [Internet]. [cited 2020 5]; http://www.ncbi.nlm.nih.gov/pubmed/31941689
Mitamura K, Shimizu H, Yamazaki M, Ichikawa M, Abe T, Yasumi Y et al (2020) Clinical evaluation of ID NOW influenza A & B 2, a rapid influenza virus detection kit using isothermal nucleic acid amplification technology—a comparison with currently available tests. J Infect Chemother 26:216–221. https://doi.org/10.1016/j.jiac.2019.08.015 [Internet]. [cited 2020 5]; https://pubmed.ncbi.nlm.nih.gov/31558351/?from_term=+ID+Now+influenza+A+%26+B+2+&from_sort=date&from_pos=3
Verbakel JY, Matheeussen V, Loens K, Kuijstermans M, Goossens H, Ieven M et al (2020) Performance and ease of use of a molecular point-of-care test for influenza A/B and RSV in patients presenting to primary care. Eur J Clin Microbiol Infect Dis. https://doi.org/10.1007/s10096-020-03860-5 [Internet]. [cited 2020 5]; http://www.ncbi.nlm.nih.gov/pubmed/32172369
Bennett S, MacLean A, Gunson R (2019) Verification of Cepheid Xpert Xpress Flu/RSV assay for use with gargle samples, sputa and endotracheal secretions. J Hosp Infect 101:114–115. https://doi.org/10.1016/J.JHIN.2018.07.016 [Internet]. [cited 2020 5]; https://www.sciencedirect.com/science/article/pii/S0195670118303803?via%3Dihub
Goldstein EJ, Gunson RN (2019) In-house validation of the cobas Liat influenza A/B and RSV assay for use with gargles, sputa and endotracheal secretions. J Hosp Infect 101:289–291. https://doi.org/10.1016/j.jhin.2018.10.025 [Internet]. [cited 2020 5]; http://www.ncbi.nlm.nih.gov/pubmed/30408505
Frazee BW, Rodríguez-Hoces de la Guardia A, Alter H, Chen CG, Fuentes EL, Holzer AK et al (2018) Accuracy and discomfort of different types of intranasal specimen collection methods for molecular influenza testing in emergency department patients. Ann Emerg Med 71:509–517.e1. https://doi.org/10.1016/j.annemergmed.2017.09.010 [Internet]. [cited 2020 5]; https://pubmed.ncbi.nlm.nih.gov/29174837/?from_single_result=Accuracy+and+Discomfort+of+Different+Types+of+Intranasal+Specimen+Collection+Methods+for+Molecular+Influenza+Testing+in+Emergency+Department+Patients
Malecki M, Lüsebrink J, Teves S, Wendel AF (2020) Pharynx gargle samples are suitable for SARS-CoV-2 diagnostic use and save personal protective equipment and swabs. Infect Control Hosp Epidemiol:1–2. https://doi.org/10.1017/ice.2020.229 [Internet]. [cited 2020 5]; http://www.ncbi.nlm.nih.gov/pubmed/32389132
Funding
This study was funded by Tosoh corporation.
Author information
Authors and Affiliations
Contributions
All authors meet the ICMJE authorship criteria.
Corresponding author
Ethics declarations
Ethical approval
This study was approved by the ethics committee of Nagasaki University Hospital (approval numbers: 17121822 and 18111919)
Consent to participate
Written informed consent for participation in and publication of this study was obtained from all participants before sample collection.
Consent for publication
Written informed consent for publication of this study was obtained from all participants before sample collection.
Competing interests
The authors declare that they have no conflict of interest.
Disclaimer
The sponsor had no control over the interpretation, writing, or publication of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
All authors meet the ICMJE authorship criteria.
Rights and permissions
About this article
Cite this article
Kaku, N., Hashiguchi, K., Akamatsu, N. et al. Evaluation of a novel rapid TRC assay for the detection of influenza using nasopharyngeal swabs and gargle samples. Eur J Clin Microbiol Infect Dis 40, 1743–1748 (2021). https://doi.org/10.1007/s10096-021-04193-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04193-7