Abstract
Bacterial vaginosis (BV) is perceived as a condition of disrupted vaginal microbiota, but remains of unknown aetiology. In this study, vaginal microbiota composition was determined in twenty-one women with BV, before and after treatment with metronidazole or clindamycin. Microbiota composition varied greatly between women and defining a (un)healthy vaginal microbiota state remains elusive, challenging BV diagnosis and treatment. While relative abundance of Lactobacillus increased after antibiotic treatment in two-third of women, its abundance was not associated with treatment outcome. Instead, remaining complaints of abnormal vaginal discharge were more common after metronidazole treatment and associated with increased relative abundance of Ureaplasma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The vaginal microbiota plays a crucial role in maintaining a healthy vaginal environment, and perturbation of this system has been implicated in disturbed vaginal health and other negative outcomes [1, 2]. The vaginal microbiota is dynamic and influenced by hormonal changes, sexual activity, and hygiene [3]. Various vaginal bacterial communities exist in healthy women, mostly dominated by Lactobacillus species, while some are being composed of anaerobes like Atopobium and Prevotella species [4]. Nevertheless, the common perception of a healthy vaginal microbiota is one dominated by one or more Lactobacillus species. As such, the switch from a Lactobacillus-dominated microbiota to a more diverse microbiota, in combination with clinical symptoms, is considered bacterial vaginosis (BV) or aerobic vaginitis, depending on colonisation by anaerobic or aerobic bacteria, respectively. Bacterial genera that are specifically associated with bacterial vaginosis are, amongst others, Gardnerella, Atopobium, Prevotella, Fusobacterium, and Dialister species [5]. Despite these associations, the aetiology of BV is unknown, and diagnosis and treatment remain elusive. While a Gram-stain evaluation according to the Nugent criteria is considered the golden standard for BV diagnosis, it is not routinely applied in a clinical setting [6]. Instead, BV diagnosis is commonly based on clinical signs and symptoms or Amsel criteria [7]. Symptoms of BV can be resolved without intervention, but metronidazole or clindamycin can be prescribed in case of persistence, even though recurrence is common [8, 9]. In our study, vaginal microbiota composition of women with clinically diagnosed BV was determined before and after antibiotic treatment and related to clinical characteristics.
Materials and methods
Prospectively, vaginal secretions and clinical data were collected from 60 premenopausal women visiting the gynaecology outpatient clinic of the Haaglanden Medical Centre (The Hague, The Netherlands) with complaints of abnormal vaginal discharge. Vaginal secretion was collected using the ESwab (Copan Diagnostics Inc., USA). BV diagnosis was based on clinical symptoms and signs, with malodorous discharge, as major criteria for diagnosis of bacterial vaginosis, followed by culturing when clinical diagnosis based on symptoms alone was uncertain. Therapy was initiated according to routine hospital practice following the European guideline and consisted of 500 mg metronidazole taken orally twice a day for 7 days, or, in case of pregnancy or lactating, 300 mg clindamycin taken orally twice a day for 7 days [10]. A follow-up visit was scheduled approximately 4 weeks after inclusion, during which vaginal swab and clinical data collection were repeated. Women who were clinically diagnosed with BV and attended the follow-up visit were selected for microbiota profiling (n = 21). Clinical data collection, Amsel criteria (vaginal pH, amine odour, wet-mount microscopy), Nugent score, and Gardnerella vaginalis culturing were performed for research purposes as previously described [11]. Detailed subject characteristics are outlined in Table 1. The Declaration of Helsinki was the guiding principle for trial execution, and the study was approved by the local ethics board (METC Zuidwest Holland, The Hague, The Netherlands). All patients provided written informed consent before participation.
Vaginal bacterial microbiota was determined by 16S rRNA gene amplicon sequencing of the V3-V4 region using the Nextera XT, MiSeq Reagent Kits v2 500 cycles, and a MiSeq desktop sequencer (Illumina, USA). Raw sequencing data are available in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under study accession PRJNA524112. Read filtering, operational taxonomic unit (OTU)-picking, and taxonomic assignment were performed using the NG-Tax 0.4 pipeline and the Silva_132_SSU Ref database [12]. Statistical analysis and data visualisation were performed in R (v3.5.1) using the packages phyloseq (v1.26.1), vegan (v2.5-4), ggplot2 (v3.1.0), DESeq2 (v1.22.2) microbiome (v1.4.2), and DirichletMultinomial (v1.24.1). For differential abundance testing by DESeq2, the OTU-table was filtered for OTUs present in less than 25% of the samples to minimise zero-variance errors and spurious significance. Permutational multivariate analysis of variance was performed using the adonis function with 999 permutations and Bray-Curtis distances to determine associations between microbiota composition and clinical variables. The Dirichlet Multinomial Mixtures method, using the Laplace equation, was applied for community typing. In this approach, samples are clustered based on microbiota profile similarity [13]. Kruskal-Wallis followed by post hoc Dunn’s testing was performed to compare Shannon diversity indices between groups.
Results and discussion
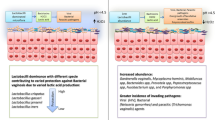
Before antibiotic treatment, genera Gardnerella, Atopobium, Prevotella, Lactobacillus, and Dialister constituted the core microbiota, and combined accounted for an average relative abundance of 71.9% (Table 2), but their abundance could vary greatly between subjects (Fig. 1a). Two community types could be identified, one driven by Gardnerella, Prevotella, Sneathia, and Atopobium (community type 1), and one driven by Lactobacillus, Gardnerella, and Atopobium (community type 2, Fig. 2a), suggesting Lactobacillus, Prevotella, and Sneathia abundances as discriminative feature of microbiota composition between patients. Bacterial diversity significantly differed between the two community types (Fig. 3a), with lower diversity in the Lactobacillus-driven community type. Microbiota composition before treatment was significantly associated with various parameters (Table 3), including the Nugent score, hormone-related variables (lactation, anticonception use), and BV symptomology (vaginal pH and amine odour).
Principal coordinate analysis and taxonomic profiles of the vaginal microbiota before (a) and after (b) antibiotic treatment. Numbers indicate individual patients. Twenty taxa with highest average relative abundance are shown; abundances of all other taxa are summed and categorised as ‘other’. For bar graphs, the subject order is matched to the subject order in the PCoA plots. Cli, clindamycin; Compl., complaints of abnormal vaginal discharge; CT1, community type one; CT2, community type 2
After treatment, bacterial diversity decreased (Fig. 3c) and the core microbiota solely consisted of Lactobacillus, constituting an average of 60.8% relative abundance (Table 2). Independent of antibiotic type (metronidazole or clindamycin), antibiotic treatment significantly decreased the relative abundance of Atopobium (Log2FoldChange = − 3.36, padj = 0.0388), while increasing Lactobacillus (Log2FoldChange = 4.04, padj = 0.0002). However, Lactobacillus remained of low abundance in one-third of the women, whose microbiota was of individual-specific composition with high abundance of either Gardnerella, Prevotella, Dialister, Escherichia-Shigella, Atopobium, or Sneathia (Fig. 1b). These microbiota compositions were also reflected by the identification of two community types: one driven by Lactobacillus, and the other driven by multiple bacterial taxa (Fig. 2b), with lower diversity in the Lactobacillus-driven community type (community type 1, Fig. 3b). Vaginal microbiota composition after antibiotic treatment was significantly associated with the Nugent score and vaginal pH (Table 3).
These findings support the current debate on the definition of a healthy vaginal microbiota [14], since Lactobacillus dominance was observed in a large proportion of women with symptoms and the opposite, dominance of anaerobes, was observed in asymptomatic women. So even in a study of small subject size, as herein, heterogeneity of vaginal bacterial communities was apparent. Vaginal health status may be associated with specific Lactobacillus species [10], which could not be defined by the method used herein. However, several kinds of microbiota composition existed in asymptomatic women, which has been previously reported [4, 11, 15]. Vaginal microbiota composition was consistently associated with the Nugent score and vaginal pH. While the Nugent score is considered the golden standard for BV diagnosis, it is rarely used in clinical setting due to resource intensiveness [6]. Determining vaginal pH is more readily applicable; however, it most certainly simply reflects the abundance of lactic acid–producing bacteria, like Lactobacillus. Nowadays, PCR-based laboratory tests would be preferred for confirmation of the diagnosis [16]. Except lactation and anticonception use, vaginal microbiota composition was not associated with patient demographics and lifestyle factors, which may be due to the relatively small subject size in combination with uniformity. It has previously been reported that host genetics, ethnicity, hormonal stage (e.g. menstruation cycle, menopause, pregnancy), sexual behaviour, and hygiene practices, amongst others factors, influence vaginal microbiota composition [17,18,19,20,21].
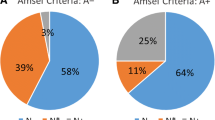
After antibiotic treatment, nine women (43%) reported remaining complaints of abnormal vaginal discharge. Persisting complaints were more prevalent in women receiving metronidazole (70%) than in those receiving clindamycin (18%), which may be a result of differences in antibiotic spectrum and underlying conditions (e.g. pregnancy). To determine the potential influence of the microbiota on clinical outcome, vaginal microbiota composition before and/or after treatment were compared between patients with and without persistent complaints. The vaginal microbiota of women with persisting complaints contained a significantly higher relative abundance of Ureaplasma (Log2FoldChange = 8.73, pajd = 0.0008), but persisting complaints could not be associated with microbiota composition before treatment. Ureaplasma is a parasitic and saprophytic bacterium belonging to the Mollicutes class and is without cell wall, which results in intrinsic resistance to cell wall–targeting antibiotics like beta-lactam and glycopeptide antibiotics [22]. Ureaplasma is intrinsically resistant to metronidazole, but usually susceptible to clindamycin [23]. While carriage of Ureaplasma in the urethra, cervix, and vagina is common and generally asymptomatic, it has previously been associated with BV recurrence [24]. Treatment outcome was not associated with the identified community types after treatment as persistent complaints were reported in 50% (7/14) and 29% (2/7) of women with vaginal microbiota composition belonging to the Lactobacillus-driven community type one or multiple species-driven community type two, respectively.
Conclusion
In conclusion, defining a (un)healthy vaginal microbiota state remains elusive, which challenges diagnosis and treatment of BV. Abnormal vaginal discharge and itching/irritation is most certainly not attributable to one or more specific bacteria, rather a disruption of the individual-specific mutualistic relationship of bacterial communities. Nevertheless, establishing universal markers for diagnosis and treatment of BV remains relevant. Herein, remaining complaints after treatment were more common in women who received metronidazole and were associated with increased relative abundance of the Ureaplasma genus, which may be considered when treatment fails.
Data availability
Raw sequencing data are available in the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under study accession PRJNA524112.
References
Greenbaum S, Greenbaum G, Moran-Gilad J, Weintraub AY (2019) Ecological dynamics of the vaginal microbiome in relation to health and disease. Am J Obstet Gynecol 220(4):324–335
Kroon SJ, Ravel J, Huston WM (2018) Cervicovaginal microbiota, women's health, and reproductive outcomes. Fertil Steril 110(3):327–336
Huang B, Fettweis JM, Brooks JP, Jefferson KK, Buck GA (2014) The changing landscape of the vaginal microbiome. Clin Lab Med 34(4):747–761
Ma B, Forney LJ, Ravel J (2012) Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol 66:371–389
Onderdonk AB, Delaney ML, Fichorova RN (2016) The human microbiome during bacterial vaginosis. Clin Microbiol Rev 29(2):223–238
Bagnall P, Rizzolo D (2017) Bacterial vaginosis: a practical review. JAAPA 30(12):15–21
Amsel R, Totten PA, Spiegel CA, Chen KC, Eschenbach D, Holmes KK (1983) Nonspecific vaginitis. Diagnostic criteria and microbial and epidemiologic associations. Am J Med 74(1):14–22
Bradshaw CS, Morton AN, Hocking J et al (2006) High recurrence rates of bacterial vaginosis over the course of 12 months after oral metronidazole therapy and factors associated with recurrence. J Infect Dis 193(11):1478–1486
Koumans EH, Markowitz LE, Hogan V, Group CBW (2002) Indications for therapy and treatment recommendations for bacterial vaginosis in nonpregnant and pregnant women: a synthesis of data. Clin Infect Dis 35(Suppl 2):S152–S172
Wang B, Xiao BB, Shang CG et al (2014) Molecular analysis of the relationship between specific vaginal bacteria and bacterial vaginosis metronidazole therapy failure. Eur J Clin Microbiol Infect Dis 33(10):1749–1756
Albert AY, Chaban B, Wagner EC et al (2015) A study of the vaginal microbiome in healthy Canadian women utilizing cpn60-based molecular profiling reveals distinct Gardnerella subgroup community state types. PLoS One 10(8):e0135620
Ducarmon QR, Hornung BVH, Geelen AR, Kuijper EJ, Zwittink RD (2020) Toward standards in clinical microbiota studies: comparison of three DNA extraction methods and two bioinformatic pipelines. mSystems 5(1)
Arumugam M, Raes J, Pelletier E et al (2011) Enterotypes of the human gut microbiome. Nature 473(7346):174–180
Reid G (2018) Is bacterial vaginosis a disease? Appl Microbiol Biotechnol 102(2):553–558
Anahtar MN, Byrne EH, Doherty KE et al (2015) Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 42(5):965–976
van den Munckhof EHA, van Sitter RL, Boers KE et al (2019) Comparison of Amsel criteria, Nugent score, culture and two CE-IVD marked quantitative real-time PCRs with microbiota analysis for the diagnosis of bacterial vaginosis. Eur J Clin Microbiol Infect Dis 38(5):959–966
Mehta SD, Nannini DR, Otieno F et al (2020) Host Genetic factors associated with vaginal microbiome composition in Kenyan women. mSystems 5(4)
Xu J, Bian G, Zheng M et al (2020) Fertility factors affect the vaginal microbiome in women of reproductive age. Am J Reprod Immunol 83(4):e13220
Fashemi B, Delaney ML, Onderdonk AB, Fichorova RN (2013) Effects of feminine hygiene products on the vaginal mucosal biome. Microb Ecol Health Dis 24
Song SD, Acharya KD, Zhu JE et al (2020) Daily vaginal microbiota fluctuations associated with natural hormonal cycle, contraceptives, diet, and exercise. mSphere 5(4)
Noyes N, Cho KC, Ravel J, Forney LJ, Abdo Z (2018) Associations between sexual habits, menstrual hygiene practices, demographics and the vaginal microbiome as revealed by Bayesian network analysis. PLoS One 13(1):e0191625
Beeton ML, Spiller OB (2017) Antibiotic resistance among Ureaplasma spp. isolates: cause for concern? J Antimicrob Chemother 72(2):330–337
Sweeney EL, Dando SJ, Kallapur SG, Knox CL (2017) The human Ureaplasma species as causative agents of chorioamnionitis. Clin Microbiol Rev 30(1):349–379
Xiao B, Wu C, Song W et al (2019) Association analysis on recurrence of bacterial vaginosis revealed microbes and clinical variables important for treatment outcome. Front Cell Infect Microbiol 9:189
Author information
Authors and Affiliations
Contributions
RZ performed microbiome analyses, interpreted data, made figures, and wrote drafts of the manuscript. EM designed the study, performed NGS, interpreted data, made the subjects table, and revised drafts of the manuscript. ML, KB, and AM designed the study and revised drafts of the manuscript. CK and EK supervised RZ and EM and interpreted data and revised drafts of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
EK is supported by an unrestricted grant from Vedanta Biosciences Inc. and has performed research for Cubist, Novartis, and Qiagen, and has participated in advisory forums of Astellas, Optimer, Actelion, Pfizer, Sanofi Pasteur, and Seres Therapeutics. These companies had no role in the research described in this manuscript. All other authors declare that they have no conflict of interest.
Ethics approval
The study was approved by the local medical ethics board (METC Zuidwest Holland, The Hague, The Netherlands).
Consent to participate
Written informed consent was obtained from all participating subjects.
Consent for publication
Written informed consent was obtained from all participating subjects.
Code availability
Raw sequencing data was analysed using free, open source software NG-Tax (http://wurssb.gitlab.io/ngtax/) and R (https://www.r-project.org/).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zwittink, R.D., van den Munckhof, E.H.A., Leverstein-van Hall, M.A. et al. The vaginal microbiota in the course of bacterial vaginosis treatment. Eur J Clin Microbiol Infect Dis 40, 651–656 (2021). https://doi.org/10.1007/s10096-020-04049-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-020-04049-6