Abstract
Foot ulcer is a common complication in diabetic subjects and infection of these wounds contributes to increased rates of morbidity and mortality. Diabetic foot infections are caused by a multitude of microbes and Staphylococcus aureus, a major nosocomial and community-associated pathogen, significantly contributes to wound infections as well. Staphylococcus aureus is also the primary pathogen commonly associated with diabetic foot osteomyelitis and can cause chronic and recurrent bone infections. The virulence capability of the pathogen and host immune factors can determine the occurrence and progression of S. aureus infection. Pathogen-related factors include complexity of bacterial structure and functional characteristics that provide metabolic and adhesive properties to overcome host immune response. Even though, virulence markers and toxins of S. aureus are broadly similar in different wound models, certain distinguishing features can be observed in diabetic foot infection. Specific clonal lineages and virulence factors such as TSST-1, leukocidins, enterotoxins, and exfoliatins play a significant role in determining wound outcomes. In this review, we describe the role of specific virulence determinants and clonal lineages of S. aureus that influence wound colonization and infection with special reference to diabetic foot infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
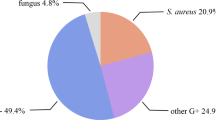
Foot ulcer is a common complication in diabetic subjects caused due to multitude of underlying risk factors including neuropathy and vascular insufficiency [1]. These open wounds favor colonization by microbes which proliferate in the wound and cause severe infection that can spread to deeper tissues thereby substantially increasing the risk of hospitalization and lower limb amputations [2]. Pathophysiology of diabetic foot infection (DFI) is complex and the wound outcome is determinant on both host factors and microbial factors including virulence [3, 4]. Diabetic foot ulcers (DFU) are quite often colonized by aerobes, anaerobes, and fungi either individually or more often as a polymicrobial community. Staphylococcus aureus, a major colonizer of DFU [5,6,7], produces abundant biofilm and thereby inhibits wound healing and exacerbates wound infection [8, 9].
S. aureus with its emerging new clones causes severe wound infection, skin and soft tissue infections (SSTI), osteomyelitis, and other unusual infections globally. Most often, S. aureus colonizes on skin or mucosal surfaces of children and HIV or diabetic patients who are more prone to S. aureus colonization [10,11,12,13]. Hospital-acquired methicillin-resistant S. aureus (MRSA) strains are largely disseminated in clinical settings and infect immunosuppressed hosts while community-associated MRSA strains can cause infections in healthy children and adults [13, 14]. Infection of mucosal surface or skin is a consequence of initial exposure eventually triggering upregulation of virulence genes [15]. S. aureus can also cause recurrent infections throughout life.
S. aureus is the predominant bacterial isolate reported from occidental countries in DFI leading to delayed wound healing. Wound adherence, persistence, and infection is enhanced by virulence factors including wide variety of enzymes and toxins elicited by S. aureus such as protease, lipases, nucleases, hyaluronidases, haemolysins (alpha, beta, gamma, and delta), and collagenase which make host tissues favorable for bacterial growth and tissue invasion. Early diagnosis and proper wound management are critical since spread of S. aureus to soft tissue and bone can significantly contribute to amputation of lower extremities [16].
Since DFU is polymicrobial nature, it is essential to consider both the microbiological and clinical features to understand microbial virulence potential of diverse microbes that cause infection and level of host susceptibility to the microbes [17, 18]. Each bacterial species differs in its virulence potential in wound environment, and it is important to evaluate the intrinsic virulence factors of isolated species to characterize and distinguish between pathogens that cause infection and colonizers [19]. In addition, it also helps to avoid misuse of antibiotics since inappropriate antibiotic usage leads to emergence of multidrug-resistant pathogens, notably MRSA. Quite often, differentiation of true infectious pathogens from colonizers is difficult especially in DFU due to the underlying risk factors of neuropathy and ischemia. In this regard, studies have been performed focusing on virulence markers and their association in wound adherence and colonization. In this review, we have focused on S. aureus virulence factors and clonal complexes commonly associated with skin and wound pathogenesis and their role in differentiating colonizing and infecting S. aureus strains in DFI.
Search strategy and selection criteria
The relevant reference articles were identified through literature search in PubMed and Web of Science databases and were restricted to those research articles published between January 1980 and March 2020. The following descriptors were used to obtain relevant references: “Staphylococcus aureus,” “ulcer,” “osteomyelitis,” “infection,” “chronic wound,” “microbiota,” “virulence,” “toxins,” “molecular methods,” “clonal complexes,” “bacterial colonization,” “antimicrobial resistance,” “adherence,” “colonization,” “genetic diversity,” “gene expression,” “host factors,” “pathogenesis,” and “biofilm” in combination with the term “Staphylococcus aureus” and “diabetic foot” or “diabetic foot osteomyelitis” and the Boolean operators AND, OR, and NOT, in addition to truncations. We have included cohort studies, cross-sectional studies, narrative reviews, and case-control studies. Only full-text articles published in English language were included. The first screening included a review of the titles of the studies. The second screening was based on the abstracts and duplicates and articles which did not meet the eligibility criteria were excluded. The final dataset included 140 full-text articles, meeting the inclusion criteria. The identified articles were reviewed and then classified based on the study objective and were then collated to understand the role of various S. aureus virulence markers and clonal complexes in wound infections.
DFU microbiome and altered physiopathology
Studies have reported the polymicrobial nature of DFU and the presence of large spectrum of microbes severely limits the use of traditional culture methods [7]. DFU is commonly colonized with aerobic Gram-positive cocci, facultative and obligate aerobic Gram-negative bacilli, obligate anaerobic bacteria [5, 20], and fungi [21]. The widespread occurrence of pathogenic and multidrug-resistant strains such as MRSA which express several virulence factors negatively influences treatment outcomes and leads to chronicity of ulcer. Screening of specific virulence genes and genotyping by multilocus sequence typing approach have shown that S. aureus isolates from monomicrobial and polymicrobial wounds differ in their clonal diversity and carriage of virulence genes [22]. Though infection in diabetic subjects by definition can include abscesses, necrotizing fasciitis, and osteomyelitis among many others, infected neuropathic diabetic foot ulcers remain the major problem [18]. Host factors such as neuropathy drastically reduce the sensory functions and pain perception causing ulceration which predisposes these wounds to severe bacterial infections [18]. Furthermore, it has been observed that early signs of infection can go undetected due to several underlying risk factors including reduced immunological functions [23], and if left untreated, the infection spreads to deeper tissues including bones. In diabetic subjects, impaired wound healing due to an increase in acute inflammatory cells, an absence of cellular growth, and decreased epidermal cell migration have been observed. In addition, the impaired host responses can shift the equilibrium from colonizers to pathogenic species leading to chronic non-healing wound ulcers.
Diabetic foot osteomyelitis
Osteomyelitis is an inflammatory condition resulting from infection of bone and is commonly missed or underdiagnosed in patients with underlying diabetic foot ulcer complications. Reports suggest that 60% of DFUs get infected and 10–15% of the infected wounds usually develop into osteomyelitis [24]. S. aureus is the primary pathogen associated with diabetic foot osteomyelitis (DFOM) and results in substantial morbidity and mortality. Studies indicate that S. aureus can form biofilms on healthy bones and infect both osteoblasts and osteoclasts while both in vivo and in vitro studies clearly show that they can also replicate and proliferate inside osteoclasts and evade destruction by immune cells [25, 26]. Interestingly, even though antibodies for various S. aureus antigens (coagulase, lukD, lukE, fibronectin-binding protein, etc.) are produced in healthy individuals, S. aureus overcomes protective immune responses and causes recurrent infections by producing pathogenic antibodies that can drastically overcome adaptive immunity [27, 28]. However, genome sequencing of two S. aureus strains collected longitudinally from a chronic osteomyelitis patient showed agrC frameshift mutations over time resulting in reduced virulence and less tissue damage [29]. Mass-based proteomics approach in a murine osteomyelitis model demonstrated that mutations in exoprotein regulatory protein saeRS and staphylococcal accessory regulator sarA attenuates virulence by downregulating virulence factor production and degradation of virulence factors respectively [30]. Víquez-Molina et al. [31] compared the prevalence of virulence genes encoding for pvl, etA, etB, and tsst in S. aureus strains in SSTI and bone infection and found no significant difference in virulence gene profiles except for higher prevalence of pvl+ strains in soft tissue infections. Even though several clonal complexes are associated with DFU colonization and infection, there are limited studies on virulence genes and clonal complexes associated with DFOM. Lattar et al. [32] performed molecular fingerprinting of S. aureus strains from patients with osteomyelitis by pulsed-field gel electrophoresis and concluded that loss of capsular polysaccharide production was the major factor associated with chronic osteomyelitis. They also showed that higher proportion of cap5 S. aureus isolates were methicillin-resistant S. aureus (MRSA) and lukS-PV/lukF-PV+ compared with cap8 isolates [32]. Senneville et al. [33] reported bone tropism of CC398 methicillin-susceptible S. aureus clone and its significance in DFOM.
Virulence factors of S. aureus
α-Toxin
In skin infections, α-toxin is considered a key virulence factor of S. aureus. This pore-forming toxin consisting primarily of beta sheets is secreted by most of the S. aureus strains as a water-soluble monomer targeting the red blood cells [34,35,36]. The gene coding for alpha toxin hla was present in S. aureus strains in all the grades of wounds in DFU though some difference was observed between MRSA and methicillin-susceptible S. aureus (MSSA) strains [37, 38].
Panton-Valentine leukocidin
Panton-Valentine leukocidin (PVL) is a potent cytotoxin that consists of two chromatographically separate protein components, namely LukS-PV (slow) and LukF-PV (fast). The active toxin causes lysis of neutrophils by forming a pore on its membrane and is associated with dermonecrosis, chronic SSTI [39, 40], recurrent mucocutaneous infections [41], and necrotizing pneumonia [42]. Further, PVL-carrying strains can cause chronic SSTI and necrotizing pneumonia in otherwise healthy individuals (Table 1). Though PVL-encoding strains are much less prevalent in community with < 10% MSSA clinical isolates found to encode pvl gene, studies indicate that isolates carrying gene coding for PVL can result in wound worsening.
Enterotoxins
S. aureus produce several exoproteins including staphylococcal enterotoxins (SEA, SEB, SECn, SED, SEE, SEG, SEH, and SEI), exfoliative toxins (ETA and ETB), and leukocidin (Fig. 1). Toxic shock syndrome toxin (TSST-1) and staphylococcal enterotoxins, collectively termed as pyrogenic toxin superantigens (PTSAgs), are known to play a significant role in proliferation of T cells irrespective of antigenic specificity. The majority of S. aureus isolates of DFU produce large number of SAgs [68], while SAg exotoxins have also been shown to contribute significantly to other major illnesses [69]. Higher number of S. aureus strains isolated from wound grades 2–4 of Wagner Classification System was shown to harbor genes encoding enterotoxins SEA and SEI than strains from grade 1 ulcer [70], making them potent markers to differentiate colonization from infection. Interestingly, S. aureus strains from DFU share more similarity with strains from atopic dermatitis and normal vaginal mucosa in their distribution and production of more types of SAgs per organism suggesting that DFU strains originated and were better adapted to skin compared with mucosae which produce fewer SAgs.
Schematic diagram illustrating major S. aureus factors associated with DFI and DFOM. (Adapted from Kong et al. [13]). ACME, arginine catabolic mobile element; agr, accessory gene regulator; Bbp, bone sialoprotein-binding protein; CC, clonal complexes; Cna, collagen adhesin; FnBP, fibronectin-binding protein; MSCRAMMs, microbial surface components recognizing adhesive matrix molecules; PMT complex, PSM transporter complex; PSM, phenol-soluble modulins; PVL, Panton-Valentine leukocidin; SAgs, super antigens; sarA, staphylococcal accessory regulator; sae, response regulator; SdrD, serine–aspartate repeat-containing protein D; SEs, staphylococcal enterotoxins; SspA, staphylococcal serine protease; SspB, cysteine protease; TSST-1, toxic shock syndrome toxin-1
Toxic shock syndrome toxin-1
Toxic shock syndrome toxin-1 (TSST-1), a 22-kD SAg, causes toxic shock syndrome. Another new member of the SAg family, SEI-X, is known to cause necrotizing pneumonia [58]. Both SEI-X and TSST-1 have potential role in DFU pathogenesis [68]. Even though the carriage of tsst-1 is low in DFU isolates, they are significantly more abundant in grade 4 ulcer than in DFOM [33].
Epidermal cell differentiation factor
The epidermal differentiation factor (EDIN) and EDIN-like factors are a family of exotoxins that specifically inhibit host protein RhoA [59], which negatively impacts host tissue by favoring bacterial dissemination and hindering complement-mediated phagocytosis. Recent findings hypothesize the role of EDIN in disseminating between tissues by hematogenous route through intracellular tunnel formation in endothelial cells named macroapertures [60, 71]. Edin-positive strains were found to be more prevalent in moderate-to-severe grade DFUs than in low-grade infection. These strains were also associated with agrI cluster and virulence markers including genes coding for hemolysin, the egc cluster of enterotoxins, lukDE, intracellular adhesion proteins (icaA, icaC, and icaD), cap5, MSCRAMM (clfA, clfB, fib, ebpS, and fnbA), and antibiotic resistance (tet and fosB). Edin-positive isolates grouped to four major clonal complexes, a singleton closely associated with CC8 (edin-A), a singleton belonging to ST152-MSSA (edin-B), CC80-MRSA (edin-B), and mostly CC25/28 MSSA (edin-A). It is also reported that grade 1 ulcer infected with edin-positive strains led to poor wound outcome [72]. While CC25/CC28-MSSA and CC80-MRSA were significantly higher in edin-positive isolates, none of them grouped to colonizing strains of CC5/CC8 [72]. Association of edin-positive strains with other virulence markers in DFU has also been reported. Thus, EDIN coding genes can be considered potent markers to categorize S. aureus strains as colonizers or infectious as well as reliable predictors of the wound outcome.
Accessory gene regulator
S. aureus pathogenicity is enhanced by quorum sensing (QS) mechanisms. Virulence factors essential for causing SSTI are regulated by accessory gene regulator (agr). Expression of several virulence determinants is known to be affected by the inhibitory activity of agr groups representing a form of bacterial interference [73]. A recent study reported that strains carrying agr were more pathogenic than those without [74].
Arginine catabolic mobile element
Arginine catabolic mobile element (ACME), a genetic island consisting of clusters of genes, confers S. aureus the ability to colonize skin. ACME is horizontally transferred from S. epidermidis, a skin commensal [75], and encodes multiple genes, among which arc (arginine deiminase system) and opp-3 (a ABC transporter) are vital in enhanced colonization. The arginine deiminase catabolizes l-arginine and by elevating skin’s pH makes it more amenable for microbial colonization [76]. Opp-3 enhances eukaryotic cell adhesion, peptide nutrient uptake, and resistance to antimicrobial peptides, thereby promoting the ability of bacteria to thrive on human skin. Thus, the acquisition of the mobile element has a potential role in disruption of skin barrier and bacterial invasion. Studies with S. aureus USA300 strain highlight the importance of ACME locus in enhancing pathogenicity and clonal dissemination. Even though, ACME has a potent role in success of USA300, its deletion has shown contradictory effect on competitive fitness in skin infection models [77, 78]. ACME speG and ACME arc genes mediate enhanced synthesis of polyamines in skin and cause clearance of S. aureus in murine skin abscess model [79]. Survival of USA300 in acidic environment is mediated by genes encoded by arc operon [79] and biofilm formation is enhanced by ACME speG-mediated polyamine tolerance [80] by upregulating genes involved in biofilm production and by increased adhesion properties, thereby favoring skin colonization, persistence, and transmission.
Microbial surface components recognizing adhesive matrix molecules
Infection of a host commences with the pathogen binding to host surface components (fibrinogen, fibronectin, and epidermal keratinocytes). A family of staphylococcal cell wall-anchored adhesins, called MSCRAMMs (microbial surface components recognizing adhesive matrix molecules), plays a significant role in aiding attachment of S. aureus virulence proteins to bone matrix and collagen [81]. In osteoblasts, MSCRAMMs play a significant role by allowing bone invasion and formation of metabolically inactive small-colony variants, which exhibit significant phenotypic and metabolic differences from regular S. aureus isolates [82,83,84,85]. However, these S. aureus variants are relatively antibiotic resistant and hinder the treatment efficacy [86, 87]. Fibronectin-binding proteins (FnBPs) are the major staphylococcal adhesins which help in colonization of human airway epithelial cells and fibroblasts and thereby establish staphylococcal infection [88]. S. aureus FnBPs also play a critical role in orthopaedic implant-associated infections, osteomyelitis, and arthritis [82].
Phenol-soluble modulins
Phenol-soluble modulins (PSMs) also play significant role in S. aureus skin infection [89]. PSMs are pore-forming toxins made up of a family of seven amphipathic α-helical peptides. Most of the S. aureus strains secrete PSMs [14] that provide them capacity to lyse human neutrophils, monocytes, erythrocytes, and osteoblasts [89] increasing tissue toxicity. Most pathogenic strains of staphylococci elicit different PSMβ peptides (PSMβ1 and 2), PSMα peptides (PSMα1–4), and a δ-toxin, thus contributing to staphylococcal pathogenesis and virulence [89]. PSMs themselves exhibit selective antimicrobial function and PSM-inspired peptides are reported to have considerable bactericidal activity against multidrug-resistant bacteria [90].
Extracellular adherence protein
Extracellular adherence protein (Eap), a 45–70-kDa protein that binds to several proteins including fibronectin, is reported to be a significant marker of impaired wound healing in mouse model [91, 92]. Eap inhibits neovascularization by hindering the inflammatory cell response near the wound area. Studies indicate that Eap interferes in ICAM-1 (intercellular adhesion molecule-1)-dependent leukocyte-endothelium interactions restricting host leukocyte recruitment, thereby aiding in persistence of S. aureus in a hostile milieu in chronic wounds [93]. In contrast, Eap does not play a major role in virulence of S. aureus in skin wound infection models as well as systemic infection models, since Eap does not contribute to bacterial adherence to proteins other than ICAM-1 [93]. However, Eap does contribute to enhanced adhesion and internalization of staphylococci by keratinocytes in a FnBP-independent manner. Eap secreted by S. aureus also significantly contributes to the internalization of other pathogenic bacteria in the wound microenvironment [94].
Biofilm factors
Biofilm production is an important strategy adopted by bacteria to colonize and infect skin tissues [95]. Though bacteria can be found in planktonic form in chronic wounds, they are most likely observed to form polymicrobial communities in biofilm matrix [96]. The presence of biofilms in non-healing wounds contributes significantly in hindering the effectiveness of antimicrobial agents and in overcoming host immunity. Bioactive compounds from biofilm communities of S. aureus and Pseudomonas aeruginosa have been shown to impair migration and proliferation of keratinocytes in chronic skin wounds and chronic tympanic membrane perforations [97]. In vitro studies also have shown that biofilm-conditioned media (BCM) from these two bacteria could inhibit cell proliferation while BCM derived from S. aureus was shown to reduce cell migration in keratinocytes and fibroblast cells in wound scratch assays [98]. Proteomic analysis of these media revealed several proteins linked to delayed wound healing including alpha hemolysin and epidermal cell differentiation inhibitor [97]. In other studies, loss of HEK cell viability by S. aureus BCM has been reported [98, 99]. HEKa cells treated with BCM showed upregulation of CXCL2, IL-8, DUSP1, and ATF3 genes which play a major role in inflammation and apoptosis [99].
Clonal complexes
Staphylococci isolated from DFU have been found to be genetically diverse, resistant to many antibiotics and harbor several virulence determinants [100]. Using multilocus sequence typing, strains of S. aureus could be grouped into clonal lineages and the major clonal lineages in humans were found to belong to clonal complex (CC)1, CC5, CC8, CC9, CC12, CC15, CC22, CC30, CC45, and CC51 [101]. In DFU, CC5 methicillin-sensitive S. aureus (CC5-MSSA), CC8-MSSA, and CC15-MSSA were considered to be colonizing strains with a favorable outcome while CC45-MSSA strains were shown to cause severe infections [37, 72, 102]. In addition, CC45 and CC30 were also considered as causative clones of severe invasive infections [103, 104]. It is believed that DFU showing worsening outcome do not colonize with CC5/CC8-MSSA strains and clonality of these strains during admission and follow-up visit remain unchanged. CC25/CC28-MSSA and CC80-MRSA strains are also considered as infecting strains in DFU as these CCs were found significantly higher in edin-positive strains (Table 2), edin gene being a predictive risk marker for worsening ulcer [72]. Even though clonal lineages found associated with humans and animals generally are different, livestock-associated CC398 (LA-CC398] strain, associated with pigs, has emerged as a major human pathogen causing severe infections [129,130,131], ventilator-associated pneumonia [132], and wound infections [133]. CC398 is significantly associated with diabetic foot osteomyelitis (DFOM) strains and helps to differentiate DFOM from SSTI—two major complications of DFU—both of which are known to carry CC45-MSSA [33]. CC398 is distinct with the presence of hemolysins, genes that code for intracellular adhesion proteins, cap5, and MSCRAMM genes including bbp, clfA and clfB [33], pvl [134], and multiple classes of antimicrobial resistance genes [135] showing potent virulence in SSTI infections in humans.
Association between presence of certain virulence genes and DFU outcome is reported in many studies. For instance, difference in the size of abscess formation in rabbit skin abscess model was attributed to different clonal lineages [136]. Different outcomes with difference in abscess diameter ranging from 5 to 7 cm (USA300, USA500, and ST80) and from 2 to 4 cm (USA400, USA1000, ST72, USA100) and almost complete absence of abscess (USA200, USA1100) were manifested by different S. aureus clonal lineages. Abscess size caused by USA300 was found to be comparable with that caused by USA100, USA200, USA400, USA1100, and ST72 strains and different from those carrying USA500, USA1000, and ST80 strains. Interestingly, though abscess formation by Panton-Valentine leukocidin (PVL)-positive USA300 and PVL-negative USA500 was comparable, the role of PVL in skin infection is thought to be limited in nature. Furthermore, neutrophil lysis activity of USA300 was shown to be significantly higher than that of other strains, and was suggested to be a major determinant of MRSA skin infection pathogenesis [136]. USA300 strains showed correlation between the expression of psmα, hla, and agr (with the exception of lukS-PV) and cause abscess, release cytokine, and lyse neutrophils lysis, whereas α-toxin and N-formylated PSMα3 peptide correlated with neutrophil lysis. Though the role of different S. aureus clonal lineages from blood stream infections is available, detailed studies on clonal types on DFI and their role in wound outcome are relatively less explored.
ST22 (CC22) is reported as a common type in DFU infections and all ST22 strains were shown to be positive for virulence factors clfa and agr I. Several less frequent clones have also been reported suggesting that diabetic patients can be an important route for dissemination of clones between hospital and community settings [100]. Sotto et al. [37] reported that foot ulcers with S. aureus strains of CC5 and CC8 showed favorable wound outcome and hypothesized that S. aureus of CC5/CC8 clones as colonizing and others as infecting clones.
Distinguishing colonization from infection in DFU
The Infectious Diseases Society of America and International Working Group on the Diabetic Foot together have established specific clinical criteria to distinguish different grades of DFI severity [137, 138]. According to this classification, grade 1 wound is considered colonized wound while grade 2 or more is considered infection. Sotto et al. [37] screened S. aureus isolates from DFU of varying grades from 1 to 4 for various virulence genes and identified several toxins including leukocidins, enterotoxins, exfoliatins, and toxic shock syndrome toxin and reported that strains from grade 1 foot ulcer to have low prevalence of virulence genes. Further, they extended their study [70] to assess clonality and carriage of 31 highly prevalent virulence-associated genes to predict the wound outcome. Among the 31 genes screened, 10 genes (sea, seb, sec, sei, sej, hlb, hlg, hlgv, cap5, and lukE) were found to be significantly associated with strains from grade 2–4 ulcers, whereas cap8 gene was associated with strains from grade 1 ulcers. None of the isolates from worsening wounds belonged to CC5 and CC8 indicating links between clonality and wound healing. However, no significant difference was found between infected and uninfected ulcers with regard to genes coding for PVL and exfoliatins [37]. But contrasting observation was found with reference to association of exfoliatins in different grades of DFU. Exfoliatin genes were found to be more likely in strains isolated from grade 4 ulcer compared with lower grades. In addition, their serotype distribution also varied with eta and etb being found very rarely (1.3%) or absent in most samples while etd (3.7%) was found in higher frequency. However, grade 1 ulcers harboring S. aureus strains carrying etd gene showed worsening wound outcome [72]. Post et al. [139] showed important differences in the presence of eta and etb gene in diabetic foot infection (eta, 13%; etb, 17%) and osteomyelitis (eta, 22% and etb, absent). One of the limitations found was the study was conducted solely on S. aureus isolates of monomicrobial wound type, while DFU is predominantly polymicrobial in nature.
Using a Caenorhabditis elegans model, Sotto et al. [70] showed that the pathogenicity of S. aureus strains in DFU grades higher than 2 were significantly more than in grade 1. Pathogenicity was assessed by the survival time of the nematode upon ingestion of S. aureus which was represented by LT50 and LT100 (time required to kill 50% and 100% of nematodes, respectively). Isolates from ulcer grades 2–4 showed LT50 < 2 days, whereas LT50 was > 3 days for isolates from grade 1 ulcer. LT50 of strains obtained from healing wounds was higher at the time of entry as well as follow-up while strains from non-healing ulcers had lower values. Messad et al. [140] identified genetic elements associated with prophage in S. aureus genome to promote colonization.
Conclusion
DFUs are extremely vulnerable to bacterial infections that can result in lower limb amputations and even death. Though from a clinician’s perspective, it is important to differentiate colonization from infection, it might prove cumbersome in DFU due to the underlying effects of neuropathy and/or ischemia. The polymicrobial community in DFI further contributes to synergistic interaction between wound pathogens and induces various virulence traits and modulates host immunity and overall wound deterioration. Prompt recognition of worsening ulcers using predictive molecular markers will hence considerably help in preventing lower limb amputations. Distribution of isolates into different clonal complexes allows comparison between colonizing and infecting strains as well as determining the origin and clonality of the strains infecting wound ulcers. Detection of specific virulence encoding genes along with clonality in different grades will help us in identifying S. aureus strains that could cause severe negative wound outcome in DFI and also to avoid misuse of antibiotic therapy in uninfected wounds.
Availability of data and material
Not applicable.
References
Lavery LA, Armstrong DG, Wunderlich RP, Mohler MJ, Wendel CS, Lipsky BA (2006) Risk factors for foot infections in individuals with diabetes. Diabetes Care 29:1288–1293
Richard J, Lavigne J, Sotto A (2012) Diabetes and foot infection: more than double trouble. Diabetes Metab Res Rev 28:46–53
Smart H, Al Ghareeb AM, Smart SA (2019) 25-Hydroxyvitamin D deficiency: impacting deep-wound infection and poor healing outcomes in patients with diabetes. Adv Skin Wound Care 32:321–328
Alvaro-Afonso FJ, Lazaro-Martinez JL, Papanas N (2018) To smoke or not to smoke: cigarettes have a negative effect on wound healing of diabetic foot ulcers. Int J Low Extrem Wounds 17:258–260
Murali TS, Kavitha S, Spoorthi J et al (2014) Characteristics of microbial drug resistance and its correlates in chronic diabetic foot ulcer infections. J Med Microbiol 63:1377–1385
Zubair M, Malik A, Ahmad J (2011) Clinico-microbiological study and antimicrobial drug resistance profile of diabetic foot infections in North India. Foot 21:6–14
Jneid J, Lavigne JP, La Scola B, Cassir N (2017) The diabetic foot microbiota: a review. Hum Microbiome J 5:1–6
Bowling FL, Jude EB, Boulton AJM (2009) MRSA and diabetic foot wounds: contaminating or infecting organisms? Curr Diabetes Rep 9:440–444
Zhao G, Usui ML, Underwood RA et al (2012) Time course study of delayed wound healing in a biofilm-challenged diabetic mouse model. Wound Repair Regen 20:342–352
Wertheim HFL, Vos MC, Ott A et al (2004) Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705
von Eiff C, Becker K, Machka K, Stammer H, Peters G (2001) Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 344:11–16
Lin J, Xu P, Peng Y et al (2017) Prevalence and characteristics of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus nasal colonization among a community-based diabetes population in Foshan, China. J Diabetes Invest 8:383–391
Kong EF, Johnson JK, Jabra-Rizk MA (2016) Community-associated methicillin-resistant Staphylococcus aureus: an enemy amidst us. PLoS Pathog 12:e1005837
Li M, Diep BA, Villaruz AE et al (2009) Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A 106:5883–5888
Novick RP (2003) Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449
Boulton AJM, Vileikyte L, Ragnarson-Tennvall G, Apelqvist J (2005) The global burden of diabetic foot disease. Lancet 366:1719–1724
Jeffcoate WJ, Lipsky BA, Berendt AR et al (2008) Unresolved issues in the management of ulcers of the foot in diabetes. Diabet Med 25:1380–1389
Lipsky BA (2004) A report from the international consensus on diagnosing and treating the infected diabetic foot. Diabetes Metab Res Rev 20:68–77
Richard JL, Sotto A, Lavigne JP (2011) New insights in diabetic foot infection. World J Diabetes 2:24–32
Smith K, Collier A, Townsend EM et al (2016) One step closer to understanding the role of bacteria in diabetic foot ulcers: characterising the microbiome of ulcers. BMC Microbiol 16:54
Chellan G, Shivaprakash S, Ramaiyar SK et al (2010) Spectrum and prevalence of fungi infecting deep tissues of lower-limb wounds in patients with type 2 diabetes. J Clin Microbiol 48:2097–2102
Shettigar K, Jain S, Bhat DV et al (2016) Virulence determinants in clinical Staphylococcus aureus from monomicrobial and polymicrobial infections of diabetic foot ulcers. J Med Microbiol 65:1392–1404
Brem H, Tomic-Canic M (2007) Cellular and molecular basis of wound healing in diabetes. J Clin Invest 117:1219–1222
Giurato L, Meloni M, Izzo V, Uccioli L (2017) Osteomyelitis in diabetic foot: a comprehensive overview. World J Diabetes 8:135–142
Krauss JL, Roper PM, Ballard A et al (2019) Staphylococcus aureus infects osteoclasts and replicates intracellularly. MBio 10:1–15
Muthukrishnan G, Masters EA, Daiss JL et al (2019) Mechanisms of immune evasion and bone tissue colonization that make Staphylococcus aureus the primary pathogen in osteomyelitis. Curr Osteoporos Rep 17:395–404
Holtfreter S, Kolata J, Broker BM (2010) Towards the immune proteome of Staphylococcus aureus— the anti-S. aureus antibody response. Int J Med Microbiol 300:176–192
Pauli NT, Kim HK, Falugi F et al (2014) Staphylococcus aureus infection induces protein A mediated immune evasion in humans. J Exp Med 211:2331–2339
Suligoy CM, Lattar SM, Llana MN et al (2018) Mutation of Agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Front Cell Infect Microbiol 2:1–9
Ramirez AM, Byrum SD, Beenken KE et al (2020) Exploiting correlations between protein abundance and the functional status of saeRS and sarA to identify virulence factors of potential importance in the pathogenesis of Staphylococcus aureus osteomyelitis. ACS Infect Dis 6:237–249
Víquez-Molina G, Aragón-Sánchez J, Pérez-Corrales C et al (2018) Virulence factor genes in Staphylococcus aureus isolated from diabetic foot soft tissue and bone infections. Int J Low Extrem Wounds 17:36–41
Lattar SM, Tuchscherr LPN, Centrón D et al (2012) Molecular fingerprinting of Staphylococcus aureus isolated from patients with osteomyelitis in Argentina and clonal distribution of the cap5(8) genes and of other selected virulence genes. Eur J Clin Microbiol Infect Dis 31:2559–2566
Senneville E, Briere M, Neut C et al (2014) First report of the predominance of clonal complex 398 Staphylococcus aureus strains in osteomyelitis complicating diabetic foot ulcers: a national French study. Clin Microbiol Infect 20:O274–O277
Otto M (2014) Staphylococcus aureus toxins. Curr Opin Microbiol 17:32–37
Xiong YQ, Willard J, Yeaman MR et al (2006) Regulation of Staphylococcus aureus α-toxin gene (hla) expression by agr, sarA and sae in vitro and in experimental infective endocarditis. J Infect Dis 194:1267–1275
Valeva A, Walev I, Pinkernell M et al (1997) Transmembrane β-barrel of staphylococcal α-toxin forms in sensitive but not in resistant cells. Proc Natl Acad Sci 94:11607–11611
Sotto A, Richard JL, Messad N et al (2012) Distinguishing colonization from infection with Staphylococcus aureus in diabetic foot ulcers with miniaturized oligonucleotide arrays: a French multicenter study. Diabetes Care 35:617–623
Djahmi N, Messad N, Nedjai S et al (2013) Molecular epidemiology of Staphylococcus aureus strains isolated from inpatients with infected diabetic foot ulcers in an Algerian University Hospital. Clin Microbiol Infect 19:E398–E404
Harch SAJ, MacMorran E, Tong SYC et al (2017) High burden of complicated skin and soft tissue infections in the indigenous population of Central Australia due to dominant Panton Valentine leucocidin clones ST93-MRSA and CC121-MSSA. BMC Infect Dis 17:1–7
Jauneikaite E, Ferguson T, Mosavie M et al (2020) Staphylococcus aureus colonization and acquisition of skin and soft tissue infection among royal marines recruits: a prospective cohort study. Clin Microbiol Infect 26:381
Balakirski G, Hischebeth G, Altengarten J et al (2020) Recurrent mucocutaneous infections caused by PVL-positive Staphylococcus aureus strains: a challenge in clinical practice. J Ger Soc Dermatol 18:315–322
Lina G, Piemont Y, Godail-Gamot F et al (1999) Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 29:1128–1132
Vazquez V, Liang X, Horndahl JK et al (2011) Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J Biol Chem 286:29797–29805
Persson L, Johansson C, Ryden C (2009) Antibodies to Staphylococcus aureus bone sialoprotein-binding protein indicate infectious osteomyelitis. Clin Vaccine Immunol 16:949–952
O’Riordan K, Lee JC (2004) Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17:218–234
Johansson A, Flock J-I, Svensson O (2001) Collagen and fibronectin binding in experimental staphylococcal osteomyelitis. Clin Orthop Relat Res 382:241–246
Patti JM, Bremell T, Krajewska-Pietrasik D et al (1994) The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun 62:152–161
Elasri MO, Thomas JR, Skinner RA et al (2002) Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone 30:275–280
Rhem MN, Lech EM, Patti JM et al (2000) The collagen-binding adhesin is a virulence factor in Staphylococcus aureus keratitis. Infect Immun 68:3776–3779
Nethercott C, Mabbett AN, Totsika M et al (2013) Molecular characterization of endocarditis-associated Staphylococcus aureus. J Clin Microbiol 51:2131–2138
Liew YK, Hamat RA, van Belkum A, Chong PP, Neela V (2015) Comparative exoproteomics and host inflammatory response in Staphylococcus aureus skin and soft tissue infections, bacteremia, and subclinical colonization. Clin Vaccine Immunol 22:593–603
Corrigan RM, Miajlovic H, Foster TJ (2009) Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9:22
Sabat A, Melles DC, Martirosian G, Grundmann H, van Belkum A, Hryniewicz W (2006) Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J Clin Microbiol 44:1135–1138
Askarian F, Ajayi C, Hanssen A-M et al (2016) The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Sci Rep 6:1–11
Trad S, Allignet J, Frangeul L et al (2004) DNA macroarray for identification and typing of Staphylococcus aureus isolates. J Clin Microbiol 42:2054–2064
Sharp JA, Echague CG, Hair PS et al (2012) Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS One 7:e38407
Tung H, Guss B, Hellman U, Persson L, Rubin K, Ryden C (2000) A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem J 345:611–619
Lemichez E, Lecuit M, Nassif X et al (2010) Breaking the wall: targeting of the endothelium by pathogenic bacteria. Nat Rev Microbiol 8:93–104
Aktories K (2011) Bacterial protein toxins that modify host regulatory GTPases. Nat Rev Microbiol 9:487–498
Boyer L, Doye A, Rolando M et al (2006) Induction of transient macroapertures in endothelial cells through RhoA inhibition by Staphylococcus aureus factors. J Cell Biol 173:809–819
Munro P, Benchetrit M, Nahori MA et al (2010) The Staphylococcus aureus epidermal cell differentiation inhibitor toxin promotes formation of infection foci in a mouse model of bacteremia. Infect Immun 78:3404–3411
Reyes-Robles T, Alonzo F, Kozhaya L, Lacy DB, Unutmaz D, Torres VJ (2013) Staphylococcus aureus leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to kill leukocytes and promote infection. Cell Host Microbe 14:453–459
Alonzo F III, Kozhaya L, Rawlings SA et al (2013) CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493:51–55
Labandeira-Rey M, Couzon F, Boisset S et al (2007) Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315:1130–1133
Spaan AN, Henry T, Van Rooijen WJM et al (2013) The staphylococcal toxin Panton-Valentine leukocidin targets human C5a receptors. Cell Host Microbe 13:584–594
Diep BA, Sensabaugh GF, Somboona NS, Carleton HA, Perdreau-Remington F (2004) Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clin Microbiol 42:2080–2084
Genestier AL, Michallet MC, Prevost G et al (2005) Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J Clin Invest 115:3117–3127
Vu BG, Stach CS, Salgado-Pabon W et al (2014) Superantigens of Staphylococcus aureus from patients with diabetic foot ulcers. J Infect Dis 210:1920–1927
Spaulding AR, Salgado-Pabón W, Kohler PL et al (2013) Staphylococcal and streptococcal superantigen exotoxins. Clin Microbiol Rev 26:422–247
Sotto A, Lina G, Richard J-L et al (2008) Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers: a new paradigm. Diabetes Care 31:2318–2324
Rolando M, Munro P, Stefani C et al (2009) Injection of Staphylococcus aureus EDIN by the Bacillus anthracis protective antigen machinery induces vascular permeability. Infect Immun 77:3596–3601
Messad N, Landraud L, Canivet B et al (2013) Distribution of edin in Staphylococcus aureus isolated from diabetic foot ulcers. Clin Microbiol Infect 19:875–880
Ji G, Beavis R, Novick RP (1997) Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030
Thompson TA, Brown PD (2017) Association between the agr locus and the presence of virulence genes and pathogenesis in Staphylococcus aureus using a Caenorhabditis elegans model. Int J Infect Dis 54:72–76
Diep BA, Gill SR, Chang RF et al (2008) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739
Diep BA, Otto M (2008) The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol 16:361–369
Diep BA, Stone GG, Basuino L et al (2008) The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis 197:1523–1530
Montgomery CP, Boyle-Vavra S, Daum RS (2009) The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background. Infect Immun 77:2650–2656
Thurlow LR, Joshi GS, Clark JR et al (2013) Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107
Planet PJ, LaRussa SJ, Dana A et al (2013) Emergence of the epidemic methicillin-resistant strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated Staphylococcus aureus adaptations for survival on skin. MBio 4:e00889–e00813
Cunningham R, Cockayne A, Humphreys H (1996) Clinical and molecular aspects of the pathogenesis of Staphylococcus aureus bone and joint infections. J Med Microbiol 44:157–164
Foster TJ (2019) The MSCRAMM family of cell-wall-anchored surface proteins of Gram-positive cocci. Trends Microbiol 27:927–941
Proctor RA, Von Eiff C, Kahl BC et al (2006) Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305
Tuchscherr L, Heitmann V, Hussain M et al (2010) Staphylococcus aureus small-colony variants are adapted phenotypes for intracellular persistence. J Infect Dis 202:1031–1040
von Eiff C, Becker K, Metze D et al (2001) Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with Darier’s disease. Clin Infect Dis 32:1643–1647
Garcia LG, Lemaire S, Kahl BC et al (2013) Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J Antimicrob Chemother 68:1455–1464
Dunyach-Remy C, Essebe CN, Sotto A, Lavigne JP (2016) Staphylococcus aureus toxins and diabetic foot ulcers: role in pathogenesis and interest in diagnosis. Toxins 8:1–20
Mongodin E, Bajolet O, Cutrona J et al (2002) Fibronectin-binding proteins of Staphylococcus aureus are involved in adherence to human airway epithelium. Infect Immun 70:620–630
Wang R, Braughton KR, Kretschmer D et al (2007) Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13:1510–1514
Zeng P, Xu C, Cheng Q et al (2019) Phenol-soluble-modulin-inspired amphipathic peptides have bactericidal activity against multidrug-resistant bacteria. ChemMedChem 14:1547–1559
Haggar A, Hussain M, Lönnies H et al (2003) Extracellular adherence protein from Staphylococcus aureus enhances internalization into eukaryotic cells. Infect Immun 71:2310–2317
Athanasopoulos AN, Economopoulou M, Orlova VV et al (2006) The extracellular adherence protein (Eap) of Staphylococcus aureus inhibits wound healing by interfering with host defense and repair mechanisms. Blood 107:2720–2727
Chavakis T, Hussain M, Kanse SM et al (2002) Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat Med 8:687–693
Bur S, Preissner KT, Herrmann M, Bischoff M (2013) The Staphylococcus aureus extracellular adherence protein promotes bacterial internalization by keratinocytes independent of fibronectin-binding proteins. J Invest Dermatol 133:2004–2012
Vlassova N, Han A, Zenilman JM et al (2011) New horizons for cutaneous microbiology: the role of biofilms in dermatological disease. Br J Dermatol 165:751–759
Cooper RA, Bjarnsholt T, Alhede M (2014) Biofilms in wounds: a review of present knowledge. J Wound Care 23:570–282
Marano RJ, Wallace HJ, Wijeratne D et al (2015) Secreted biofilm factors adversely affect cellular wound healing responses in vitro. Sci Rep 5:13296
Kirker KR, Secor PR, James GA, Fleckman P, Olerud JE, Stewart PS (2009) Loss of viability and induction of apoptosis in human keratinocytes exposed to Staphylococcus aureus biofilms in vitro. Wound Repair Regen 17:690–699
Tankersley A, Frank MB, Bebak M et al (2014) Early effects of Staphylococcus aureus biofilm secreted products on inflammatory responses of human epithelial keratinocytes. J Inflamm (Lond) 11:17
Mottola C, Semedo-Lemsaddek T, Mendes JJ et al (2016) Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J Biomed Sci 23:1
Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T et al (2003) How clonal is Staphylococcus aureus? J Bacteriol 185:3307–3316
Kolawole DO, Adeyanju A, Schaumburg F et al (2013) Characterization of colonizing Staphylococcus aureus isolated from surgical wards’ patients in a Nigerian university hospital. PLoS One 8:e68721
Wehrhahn MC, Robinson JO, Pascoe EM et al (2012) Illness severity in community-onset invasive Staphylococcus aureus infection and the presence of virulence genes. J Infect Dis 205:1840–1848
Xiong YQ, Fowler VG, Yeaman MR, Perdreau-Remington F, Kreiswirth BN, Bayer AS (2009) Phenotypic and genotypic characteristics of persistent methicillin-resistant Staphylococcus aureus bacteremia in vitro and in an experimental endocarditis model. J Infect Dis 199:201–208
Egyir B, Guardabassi L, Sorum M et al (2014) Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from healthcare institutions in Ghana. PLoS One 9:e89716
Menegotto F, Gonzalez-Cabrero S, Cubero A et al (2012) Clonal nature and diversity of resistance, toxins and adhesins genes of meticillin-resistant Staphylococcus aureus collected in a Spanish hospital. Infect Genet Evol 12:1751–1758
Xiao M, Zhao R, Zhang Q et al (2016) Genotypic diversity of Staphylococcus aureus α-hemolysin gene (hla) and its association with clonal background: implications for vaccine development. PLoS One 11:e0149112
Ruzickova V, Pantucek R, Petras P, Machova I, Kostylkova K, Doskar J (2012) Major clonal lineages in impetigo Staphylococcus aureus strains isolated in Czech and Slovak maternity hospitals. Int J Med Microbiol 302:237–241
Shittu AO, Oyedara O, Okon K et al (2015) An assessment on DNA microarray and sequence-based methods for the characterization of methicillin-susceptible Staphylococcus aureus from Nigeria. Front Microbiol 6:1–9
Lozano C, Porres-Osante N, Crettaz J et al (2013) Changes in genetic lineages, resistance, and virulence in clinical methicillin-resistant Staphylococcus aureus in a Spanish hospital. J Infect Chemother 19:233–242
Tokajian ST, Khalil PA, Jabbour D et al (2010) Molecular characterization of Staphylococcus aureus in Lebanon. Epidemiol Infect 138:707–712
Luedicke C, Slickers P, Ehricht R, Monecke S (2010) Molecular fingerprinting of Staphylococcus aureus from bone and joint infections. Eur J Clin Microbiol Infect Dis 29:457–463
van Trijp MJCA, Melles DC, Snijders SV et al (2010) Genotypes, superantigen gene profiles, and presence of exfoliative toxin genes in clinical methicillin-susceptible Staphylococcus aureus isolates. Diagn Microbiol Infect Dis 66:222–224
Peacock SJ, Moore CE, Justice A et al (2002) Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun 70:4987–4996
Li S, Sun J, Zhang J et al (2014) Comparative analysis of the virulence characteristics of epidemic methicillin–resistant Staphylococcus aureus (MRSA) strains isolated from Chinese children: ST59 MRSA highly expresses core gene–encoded toxin. Apmis 122:101–114
Croes S, Deurenberg RH, Boumans MLL, Beisser PS, Neef C, Stobberingh EE (2009) Staphylococcus aureus biofilm formation at the physiologic glucose concentration depends on the S. aureus lineage. BMC Microbiol 9:1–9
Blomfeldt A, Eskesen AN, Aamot HV, Leegaard TM, Bjornholt JV (2016) Population-based epidemiology of Staphylococcus aureus bloodstream infection: clonal complex 30 genotype is associated with mortality. Eur J Clin Microbiol Infect Dis 35:803–813
Cheung GYC, Kretschmer D, Duong AC et al (2014) Production of an attenuated phenol-soluble modulin variant unique to the MRSA clonal complex 30 increases severity of bloodstream infection. PLoS Pathog 10:e1004298
Rieg S, Jonas D, Kaasch AJ et al (2013) Microarray-based genotyping and clinical outcomes of Staphylococcus aureus bloodstream infection: an exploratory study. PLoS One 8:e71259
Wu D, Li X, Yang Y et al (2011) Superantigen gene profiles and presence of exfoliative toxin genes in community-acquired meticillin-resistant Staphylococcus aureus isolated from Chinese children. J Med Microbiol 60:35–45
Ghasemzadeh-Moghaddam H, Ghaznavi-Rad E, Sekawi Z et al (2011) Methicillin-susceptible Staphylococcus aureus from clinical and community sources are genetically diverse. Int J Med Microbiol 301:347–353
Yamaguchi T, Okamura S, Miura Y, Koyama S, Yanagisawa H, Matsumoto T (2015) Molecular characterization of community-associated methicillin-resistant Staphylococcus aureus isolated from skin and pus samples of outpatients in Japan. Microb Drug Resist 21:441–447
Stieber B, Monecke S, Muller E, Baier V, Coombs GW, Ehricht R (2014) Development and usage of protein microarrays for the quantitative measurement of Panton-Valentine leukocidin. Mol Cell Probes 28:123–132
Bennett CM, Coombs GW, Wood GM et al (2014) Community-onset Staphylococcus aureus infections presenting to general practices in South-eastern Australia. Epidemiol Infect 142:501–511
Shore AC, Tecklenborg SC, Brennan GI, Ehricht R, Monecke S, Coleman DC (2014) Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. J Clin Microbiol 52:859–870
Chen L, Mediavilla JR, Smyth DS et al (2010) Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob Agents Chemother 54:3347–3354
Deurenberg RH, Rijnders MIA, Sebastian S, Welling MA, Beisser PS, Stobberingh EE (2009) The Staphylococcus aureus lineage-specific markers collagen adhesin and toxic shock syndrome toxin 1 distinguish multilocus sequence typing clonal complexes within spa clonal complexes. Diagn Microbiol Infect Dis 65:116–122
Monecke S, Müller E, Büchler J, Stieber B, Ehricht R (2014) Staphylococcus aureus in vitro secretion of alpha toxin (hla) correlates with the affiliation to clonal complexes. PLoS One 9:e100427
Schmidt T, Zündorf J, Grüger T et al (2013) Phenotyping of Staphylococcus aureus reveals a new virulent ST398 lineage. Clin Microbiol Infect 19:279–285
Fitzgerald JR (2012) Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol 20:192–198
McCarthy AJ, Van Wamel W, Vandendriessche S et al (2012) Staphylococcus aureus CC398 clade associated with human-to-human transmission. Appl Environ Microbiol 78:8845–8848
Witte W, Strommenger B, Stanek C, Cuny C (2007) Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, central Europe. Emerg Infect Dis 13:255–258
Kock R, Schaumburg F, Mellmann A et al (2013) Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS One 8:e55040
Zhao C, Liu Y, Zhao M et al (2012) Characterization of community acquired Staphylococcus aureus associated with skin and soft tissue infection in Beijing: high prevalence of PVL+ ST398. PLoS One 7:e38577
Price LB, Stegger M, Hasman H et al (2012) Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3:1–6
Li M, Cheung GYC, Hu J et al (2010) Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis 202:1866–1876
Lipsky BA, Aragon-Sanchez J, Embil J et al (2016) International Working Group on the Diabetic Foot (IWGDF). IWGDF guidance on the diagnosis and management of foot infections in persons with diabetes. Diabetes Metab Res Rev 32:45–74
Lipsky BA, Senneville É, Abbas ZG et al (2020) Guidelines on the diagnosis and treatment of foot infection in persons with diabetes (IWGDF 2019 update). Diabetes Metab Res Rev 36:1–24
Post V, Wahl P, Uckay I et al (2014) Phenotypic and genotypic characterisation of Staphylococcus aureus causing musculoskeletal infections. Int J Med Microbiol 304:565–576
Messad N, Prasjnar TK, Lina G et al (2015) Existence of a colonizing Staphylococcus aureus strain isolated in diabetic foot ulcers. Diabetes 64:2991–2995
Acknowledgments
The authors thank TIFAC-CORE in Pharmacogenomics for funding, DST-FIST, Government of India for facilities, Manipal Academy of Higher Education for the support and funding. KS thanks DST – INSPIRE, Government of India for fellowship. TSM thanks Ms. Apoorva Jnana for help and Dr. Satyamoorthy for support and encouragement.
Code availability
Not applicable
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal.
Author information
Authors and Affiliations
Contributions
Both the authors contributed to the idea of the article. Literature search and data analysis were performed by Kavitha. First draft was written by Kavitha and critically revised by Murali. Both the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
Not applicable
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shettigar, K., Murali, T.S. Virulence factors and clonal diversity of Staphylococcus aureus in colonization and wound infection with emphasis on diabetic foot infection. Eur J Clin Microbiol Infect Dis 39, 2235–2246 (2020). https://doi.org/10.1007/s10096-020-03984-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-020-03984-8