Abstract
Acinetobacter baumannii is an important cause of multidrug-resistant hospital acquired infections in the world. Here, we investigate the presence of NDM-1 and other carbapenemases among carbapenem-resistant A. baumannii isolated between August 2010 and December 2014 from three large hospitals in Hanoi, Vietnam. We identified 23/582 isolates (4 %) (11 from hospital A, five from hospital B, and seven from hospital C) that were NDM-1 positive, and among them 18 carried additional carbapenemase genes, including seven isolates carrying NDM-1, IMP-1, and OXA-58 with high MICs for carbapenems. Genotyping indicated that NDM-1 carrying A. baumannii have expanded clonally in these hospitals. Five new STs (ST1135, ST1136, ST1137, ST1138, and ST1139) were identified. One isolate carried NDM-1 on a plasmid belonging to the N-repA replicon type; no NDM-1-positive plasmids were identified in the other isolates. We have shown the extent of the carbapenem resistance and the local clonal spread of A. baumannii carrying NDM-1 in these hospitals; coexistence of NDM-1 and IMP-1 is reported for the first time from Vietnam here, and this will further seriously limit future therapeutic options.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance (AMR) has taken centre stage as a global health issue demanding public attention and commanding resources to understand where the international community will be placed in in 2020/2025 [1]. AMR is now deemed to be the biggest global threat facing humanity in the twenty-first century. The World Health Organisation recently published a mapping exercise on regional capacity, and South-East Asia lacked many basic reporting structures on AMR [2]. The problem of antibiotic resistance has recently reached a climax with the discovery of New Delhi metallo-beta-lactamase 1 (NDM-1) and MCR-1, plasmid-borne genes that confer resistance to the last-resort antibiotics carbapenems and colistin respectively [3, 4].
Many would argue that it was the advent of the NDM-1 metallo-beta-lactamase resistance mechanism appearing in Europe and being strongly linked to South Asia (mainly India and Pakistan) that became the universal game changer for the international importance of AMR [5]. It is now recognised that this is as much a public health issue (unabated dissemination in the community) as a hospital-based issue [6]. The NDM-1 gene encodes an enzyme that hydrolyses and inactivates all beta-lactam antibiotics including carbapenems, except for aztreonam, and thus induces resistance to carbapenems [3]. This gene has been identified on a variety of plasmids in a large number of Gram-negative pathogens and reported from most continents around the globe, especially from South-East Asia [6, 7].
Acinetobacter baumannii is an opportunistic pathogen in humans, and is an increasing cause of drug-resistant hospital-acquired infections across the world [8, 9]. A. baumannii is a strictly aerobic, non-motile, Gram-negative coccobacillus belonging to the Acinetobacter calcoaceticus–baumannii complex, within the family Moraxellaceae of the order Gammaproteobacteriae. In contrast to other Acinetobacter species, A. baumannii is more common in hospital environments and is capable of surviving on dry surfaces for months [10]. Resistance to carbapenems in A. baumannii is becoming common across all continents [9, 11]. The resistance is mainly mediated by OXA-type serine beta-lactamases, and IMP-type and VIM-type metallo-beta-lactamases [9].
NDM-1 now has been found in several different species of Enterobacteriaceae. A. baumannii carrying NDM-1 have been reported from clinical and environmental isolates in several countries [12–16].
In Vietnam, antibiotic resistance rates have been reportedly high. A combination of a high infectious diseases burden, unrestricted access to antibiotics, and poor infection control measures contribute to the emergence of antibiotic resistance in Vietnam [17, 18]. Enterobacteriaceae carrying NDM-1 have been reported from Vietnamese patients, healthy volunteers, and the environment [19–21]. A. baumannii is one of three (the others are Pseudomonas aeruginosa and Klebsiella pneumoniae) most common pathogens of hospital acquired infections (HAIs), accounting for 23.8 % of all HAIs causes in Vietnam, and almost 50 % of these were resistant to carbapenems [22]. The carbapenem resistance was caused by OXA-23 and was found in clinical and environmental isolates [7, 23]. Recently, NDM-1-producing A. baumannii was described from a hospital in Ho Chi Minh City in southern Vietnam [24]. Here, we collected isolates from three hospitals in Hanoi to detect carbapenem-resistant A. baumannii containing the NDM-1 gene and to determine the relationship of NDM-1-carrying isolates between these hospitals.
Materials and methods
Study sites
Between August 2010 and December 2014, we prospectively collected data and carbapenem-resistant A. baumannii isolates from three large hospitals: Saint Paul (A), Thanh Nhan (B) and Vietduc (C) in Hanoi, Vietnam.
Demographic and basic clinical information were collected from patients from whom carbapenem-resistant bacteria were cultured, including: age, gender, date of admission, clinical diagnosis, origin of collected sample, isolated bacterial strains, and date of sample collection. All of clinical isolates used in this study were obtained during standard patient care. Treatment and clinical outcome data were not available for this study. Isolates were sent to the National Institute of Hygiene and Epidemiology (NIHE) for further characterisation.
Bacterial identification, susceptibility testing and detection of resistance genes
The microbiology laboratories of each hospital performed culture and species identification using commercial biochemical testing kits (API20E and API20NE, Biomérieux, Marcy l’Étoile, France). Minimum inhibitory concentrations (MICs) for carbapenem were performed by agar dilution according to CLSI 2012 guidelines and European Committee on Antimicrobial Susceptibility testing (EUCAST) breakpoints for colistin [25, 26]. The antibiotics used for MIC were imipenem (IMP), meropenem (MEM), cefotaxime (CTX), ceftazidime (CAZ), ciprofloxacin (CIP), and colistin (CS) (Sigma–Aldrich). The MBL E-test (Biomérieux) was used to screen for metallo-beta-lactamase production. NDM-1 was detected by polymerase chain reaction (PCR) using specific primers [3], other carbapenemases genes (KPC, IMP, VIM, SIM), and OXA genes as described elsewhere [27–30]. Resulting amplicons were sequenced using conventional sequencing.
PFGE
Pulsed-field gel electrophoresis (PFGE) was carried following digestion with ApaI (Roche Diagnostic, Mannheim, Germany). Salmonella braenderup H9812 digested with XbaI served as reference molecular weight marker. DNA fragments were separated on a CHEF-DR III apparatus (Bio-Rad, Hercules, CA, USA) for 20 h at 6 V/cm at 14 °C with an initial pulse time of 0.5 s and final pulse time of 30s, as described elsewhere [5].
Multilocus sequence typing (MLST)
MLST using the Oxford scheme was performed according to the protocol described on the MLST website (http://pubmlst.org/abaumannii/). Seven housekeeping genes were amplified by PCR, and sequenced and compared with the sequences submitted to the MLST database websites to determine Sequence Types (STs).
Plasmid characterization
DNA plugs of the isolates harbouring bla NDM-1 were treated with restriction enzyme S1 (Invitrogen, Abingdon, UK) with Salmonella braenderup H9812 digested with XbaI as reference molecular weight marker and separated by PFGE. The Biometra–Analytik system (Jena, Germany) was used to transfer DNA fragments from the gel to nylon-membranes. Fragments were hybridised with NDM-1 probe labeled in HL-2000 HybriLinker (UPV, Germany). Enterobacter cloacae carrying the NDM-1 plasmid was used as positive control [7]. Plasmids hybridising with the NDM-1 probe were cut from the gel, purified, and typed as described elsewhere [31]
Conjugational transfer of NDM-1 plasmids to the laboratory strain E. coli J53 was done on Luria–Bertani broth without selection. After 16 h, the mixed culture was centrifuged, suspended in saline, and plated onto MacConkey agar containing sodium azide (100 mg/l) and meropenem (0.5 mg/l). Transconjugants were confirmed to have NDM-1 by PCR analysis. Plasmids were subsequently isolated and typed as described elsewhere [31].
Statistical methods
PFGE data were analysed using BioNumerics version 6.5 (Applied Maths, USA). Isolates and patient data were analysed in MS Excel 2010 (Microsoft Corp., USA) using descriptive statistics as appropriate.
Ethics statement
Ethical approval was obtained from the Ethical Committee of NIHE in 2010. All patient data were anonymised.
Results
Between August 2010 and December 2014 1783 carbapenem-resistant Gram-negative bacteria were collected, 582 (32.6 %) of which were A. baumannii. A baumannii isolates were cultured from all three participating hospitals: 34 isolates in 2010, 220 in 2011, 279 in 2012, 47 in 2013, and two isolates in 2014. Fifty-five isolates were from hospital A, 106 from hospital B, and 421 from hospital C. Among these 582, A. baumannii isolates, 23 (4.0 %) were confirmed to be NDM-1-positive: 11 from hospital A, five from hospital B, and seven from hospital C (Table 2). In this study, we also investigated the presence of other carbapenem resistance genes. Among 559 carbapenem-resistant A. baumannii isolates negative to NDM-1, 550 (98.4 %) were positive for OXA-23, and two (0.35 %) isolates were positive for OXA-58. Two A. baumannii isolates were positive for IMP-1. The remaining A. baumannii isolates were negative for KPC, VIM, SIM, and OXA-48 genes (Table 1).
The median age of the patients with NDM-1-positive isolates was 26 (range: 1 to 77 years). Eight patients were under 1 year of age. Among NDM-1-positive patients, 20 were male and three were female. Seventeen strains were isolated from endotracheal aspirates, three from blood, two from urine and one from pus.
Interestingly, 18/23 NDM-1-producing A. baumannii carried additional carbapenemase genes: NDM-1, IMP-1, and OXA-58 were found in seven isolates from 2010–2011 in hospital A. One isolate in hospital B in 2011 was positive for NDM-1, OXA-23, and OXA-58. Eight isolates were positive for NDM-1 and OXA-58 (three in hospital A, one in hospital B, and four in hospital C), and two isolates were carrying both NDM-1 and OXA-23 in hospital A (2014) and C (2013) (Table 2).
All carbapenem-resistant isolates were found to be susceptible to colistin, and 13/23 isolates (57 %) remained susceptible to ciprofloxacin. All isolates were resistant to ceftriaxone and ceftazidime. For carbapenems, one A. baumannii isolate from blood in hospital B was susceptible to meropenem (0.5 mg/l) but had reduced susceptibility to imipenem (2 mg/l), and 22/23 isolates were resistant to both imipenem and meropenem (8–256 mg/l). A significantly higher level of carbapenem resistance was detected in 7 A. baumannii with co-existent NDM-1, IMP-1, and OXA-58 (MIC imipenem = 128-256 mg/l, median 256, and MIC meropenem = 64–128 mg/l, median 128) when compared with other NDM-1-producing A. baumannii (MIC = 8–128 mg/l for both imipenem and meropenem, median 32 and 16 respectively, p = 0.001) (Table 2).
In PFGE analysis of the 23 bla NDM-1-positive isolates recovered in this study, four PFGE clusters were observed (Fig. 1). Cluster I contains three isolates from hospital B in 2013, cluster II contains two isolates in 2011 from hospital B and C, cluster III contains eight isolates (seven in 2011 and one isolate in 2012) in hospital A, and cluster IV contains two isolates from hospital C. These results indicate that NDM-1-carrying A. baumannii has expanded clonally in these hospitals and has the potential for intra-hospital patient-to-patient spread.
The A. baumannii isolates producing NDM-1 belonged to ST91, ST208, ST302, ST493, ST593, ST655, ST861, ST1097, ST1261, and to five new STs: ST1135, ST1136, ST1137, ST1138, and ST1139. ST1139 belongs to international clone CC92, and was found in the ICU of hospital C in 2010 (Fig. 1 and Table 2). Six of the A. baumannii isolates producing NDM-1 from bronchial fluid of patients in hospital A belonged to the new ST1135, and two new ST302 were found in hospital B and C. We have also analysed a selection (8/559) of carbapenem-resistant A. baumannii negative to NDM-1, and these isolates belonged to ST91, ST92, ST109, ST195, ST495, ST620, and ST1140.
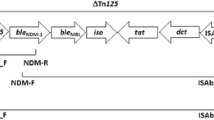
Southern blotting only detected NDM-1 in one plasmid belonging to the N-repA replicon type, isolated from A. baumannii strain 1146 from blood of a patient in the ICU of hospital B (Fig. 2). Conjugation assay was performed, but we found no transconjugant strains. There was lack of plasmid in 22/23 positive NDM-1, suggesting that spread is mainly clonal and not through horizontal gene transfer.
S1-PFGE and Southern blotting of plasmids carrying NDM-1 from clinical isolates. Pulse-field gel of S1 nuclease-treated plasmid DNA of selected A. baumannii from clinical isolates in three hospitals, stained with ethidium bromide. The molecular weight marker is Salmonella braenderup H9812 cut with XbaI (a). Autodiagram of gel A showing plasmids carrying NDM-1 (b). Enterobacter cloacae positive NDM-1 plasmid from Vietduc hospital
Discussion
Since the first description of NDM-1 [3], the rapid dissemination of this gene among Enterobacteriaceae has been reported from many countries around the world [5, 6]. However, NDM-1-production has also been found in other Gram-negative bacteria, such as A. baumannii [12–16]. In Vietnam, NDM-1-producing Enterobacteriaceae were commonly isolated from patients admitted to a Vietnamese surgical hospital, and from the environment [19, 20], and recently two NDM-1 producing A. baumannii were described from a hospital in Ho Chi Minh City in southern Vietnam [24]. NDM genes have been described in A. baumannii from other countries from 2010 onwards, and NDM associated with the Tn125 transposon has been hypothesised to originate from A. baumannii isolates in North Africa prior to transfer to Enterobacteriaceae [8, 32].
Here, we report for the first time the isolation of NDM-1-producing A. baumannii from patients in northern Vietnam in 23/582 isolates (4 %) collected between 2010 and 2014, with the first isolate being detected in 2010. As is commonly found in other countries, the majority of NDM-1 was not detected on plasmids and presumed to be integron-associated or chromosomal [33]. Also, NDM-1 was found to co-exist with other carbapenemases including IMP-1 reported in Vietnam for the first time, and OXA-58. The contribution of either of these genes to the carbapenem resistance is unclear, but MIC of isolates co-carrying IMP-1 was significantly higher.
Low level carbapenem resistance in A. baumannii is often mediated through OXA-51 like enzymes belonging to the class D ß-lactamase family, which exhibit weak carbapenemase activity and are labeled CHDL for carbapenem-hydrolysing class D ß-lactamases. OXA-23 is the most commonly identified CHDL world-wide in A. baumannii, and was also found in 550/559 NDM-1-negative carbapenem-resistant isolates and 3/23 NDM-1 positive isolates. OXA-58 was detected in 2/559 NDM-1-negative and 16/23 NDM-1-positive isolates. True carbapenemase activity (metallo-ß-lactamases, class B ß-lactamases) in A. baumannii can be mediated by IMP, VIM, SIM-1, NDM, and other genes. Here, we only detected IMP-1 in 2/550 NDM-1-negative and 7/23 NDM-1-positive isolates. This distribution of resistance genes suggests different phylogenies of the NDM-1-positive and -negative isolates, which was confirmed by MLST. There was limited overlap with STs described in a large Asian surveillance project [34]. MLST further showed several new sequence types and clustering of isolates suggestive of local evolution, clonal expansion, and within-hospital spread.
This study shows the proportion of carbapenem-resistant A. baumannii isolates carrying carbapenemase gene in three large hospitals in Ha Noi. There are limitations to the results of this study that are caused by the limited amount of metadata that were collected, and by the fact that we only analysed carbapenem-resistant isolates that were found in the laboratory among samples sent in to the laboratory by treating physicians. Therefore, we cannot draw conclusions whether isolates were likely to be community- or hospital-acquired, and whether they were causing illness or commensal, whether clustering is due to the hospital environment serving as a reservoir or patient-to-patient transmission, what the percentage of total isolates of A. baumannii with carbapenem resistance is, and other epidemiologic analyses requiring hospital and laboratory denominators.
Conclusion
In this study, we show that NDM-1 has already been present in A. baumannii in northern Vietnam since 2010. The extent of the carbapenem resistance and the local clonal spread of A. baumannii carrying NDM-1 in these hospitals, as well as coexistence of NDM-1 and IMP-1, is reported for the first time from Vietnam here, and this will further seriously limit future therapeutic options.
References
O’Neill J (2014) Review on Antimicrobial Resistance. Antimicrobial Resistance: tackling a crisis for the health and wealth of nations
World Health Organisation (2014) Antimicrobial Resistance: Global Report on surveillance
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K et al (2009) Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53(12):5046–5054
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J et al (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16(2):161–168
Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R et al (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10(9):597–602
Poirel L, Hombrouck-Alet C, Freneaux C, Bernabeu S, Nordmann P (2010) Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect Dis 10(12):832
Tran HH, Ehsani S, Shibayama K, Matsui M, Suzuki S, Nguyen MB et al (2015) Common isolation of New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae in a large surgical hospital in Vietnam. Eur J Clin Microbiol Infect Dis 34(6):1247–1254
Poirel L, Bonnin RA, Nordmann P (2011) Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 63(12):1061–1067
Poirel L, Nordmann P (2006) Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 12(9):826–836
Peleg AY, Seifert H, Paterson DL (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21(3):538–582
Al-Sultan AA, Evans BA, Aboulmagd E, Al-Qahtani AA, Bohol MF, Al-Ahdal MN et al (2015) Dissemination of multiple carbapenem-resistant clones of Acinetobacter baumannii in the Eastern District of Saudi Arabia. Front Microbiol 6:634
Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y (2012) Prevalence of New Delhi metallo-beta-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune. India Int J Antimicrob Agents 39(3):265–266
Jones LS, Toleman MA, Weeks JL, Howe RA, Walsh TR, Kumarasamy KK (2014) Plasmid carriage of bla NDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob Agents Chemother 58(7):4211–4213
Krizova L, Bonnin RA, Nordmann P, Nemec A, Poirel L (2012) Characterization of a multidrug-resistant Acinetobacter baumannii strain carrying the blaNDM-1 and blaOXA-23 carbapenemase genes from the Czech Republic. J Antimicrob Chemother 67(6):1550–1552
Nemec A, Krizova L (2012) Carbapenem-resistant Acinetobacter baumannii carrying the NDM-1 gene, Czech Republic, 2011. Euro Surveill 17(11):pii: 20121
Zhang C, Qiu S, Wang Y, Qi L, Hao R, Liu X et al (2014) Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One 8(6), e64857
Nguyen KV, Thi Do NT, Chandna A, Nguyen TV, Pham CV, Doan PM et al (2013) Antibiotic use and resistance in emerging economies: a situation analysis for Viet Nam. BMC Public Health 13:1158
TT N d, Chuc NT, Hoa NP, Hoa NQ, Nguyen NT, Loan HT et al (2014) Antibiotic sales in rural and urban pharmacies in northern Vietnam: an observational study. BMC Pharmacol Toxicol 15(1):6
Hoang TH, Wertheim H, Minh NB, Duong TN, Anh DD, Phuong TT et al (2013) Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae strains containing New Delhi metallo-beta-lactamase isolated from two patients in Vietnam. J Clin Microbiol 51(1):373–374
Isozumi R, Yoshimatsu K, Yamashiro T, Hasebe F, Nguyen BM, Ngo TC et al (2012) bla(NDM-1)-positive Klebsiella pneumoniae from environment, Vietnam. Emerg Infect Dis 18(8):1383–1385
Dao TT, Liebenthal D, Tran TK, Ngoc Thi Vu B, Ngoc Thi Nguyen D, Thi Tran HK et al (2014) Klebsiella pneumoniae oropharyngeal carriage in rural and urban Vietnam and the effect of alcohol consumption. PLoS One 9(3), e91999
Global Antibiotic Resistance Partnership (2010) Situation Analysis: Antibiotic Use and Resistance in Vietnam
Tada T, Miyoshi-Akiyama T, Kato Y, Ohmagari N, Takeshita N, Hung NV et al (2013) Emergence of 16S rRNA methylase-producing acinetobacter baumannii and pseudomonas aeruginosa isolates in hospitals in Vietnam. BMC Infect Dis 13:251
Tada T, Miyoshi-Akiyama T, Shimada K, Nga TT, le Thu TA, Son NT et al (2015) Dissemination of clonal complex 2 Acinetobacter baumannii strains co-producing carbapenemases and 16S rRNA methylase ArmA in Vietnam. BMC Infect Dis 15:433
Clinical and Laboratory Standards Institute (2012) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard-Ninth Edition (M07-A9)
European Committee on Antimicrobial Susceptibility Testing (2014) Clinical Breakpoints — bacteria v4.0
Bratu S, Mooty M, Nichani S, Landman D, Gullans C, Pettinato B et al (2005) Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob Agents Chemother 49(7):3018–3020
Ellington MJ, Kistler J, Livermore DM, Woodford N (2007) Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother 59(2):321–322
Poirel L, Heritier C, Tolun V, Nordmann P (2004) Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48(1):15–22
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S et al (2006) Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 27(4):351–353
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63(3):219–228
Karthikeyan K, Thirunarayan MA, Krishnan P (2010) Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother 65(10):2253–2254
Potron A, Poirel L, Nordmann P (2015) Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45(6):568–585
Kim DH, Choi JY, Kim HW, Kim SH, Chung DR, Peck KR et al (2013) Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob Agents Chemother 57(11):5239–5246
Acknowledgments
This work was supported by a grant from the National Foundation for Science and Technology Development (NAFOSTED; MS106.03-2012.44), Vietnam; the Wellcome Trust, UK; and Grant-in-aid of Ministry of Health, Labor and Welfare; the Government of Japan (H23-Shinkou- shitei-020).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None declared.
Ethical approval
Ethical approval was obtained from the Ethical Committee of NIHE in 2010.
Informed consent
All patient were taken Informed consent
Additional information
One sentence summary
We found a high rate of isolation of carbapenem-resistant A. baumannii, and report co-existence of NDM-1 and IMP-1 for the first time from Vietnam, requiring urgent control efforts.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Tran, D.N., Tran, H.H., Matsui, M. et al. Emergence of New Delhi metallo-beta-lactamase 1 and other carbapenemase-producing Acinetobacter calcoaceticus-baumannii complex among patients in hospitals in Ha Noi, Viet Nam. Eur J Clin Microbiol Infect Dis 36, 219–225 (2017). https://doi.org/10.1007/s10096-016-2784-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2784-8