Abstract
Comparatively few studies have been published describing Staphylococcus aureus/MRSA epidemiology in Central Asia including Pakistan. Here, we report the genotyping of Staphylococcus aureus strains (that include both methicillin-susceptible and methicillin-resistant Staphylococcus aureus) from community- and hospital-acquired skin and soft-tissue infections in a tertiary care hospital in the Malakand district of the Khyber Pakhtunkhwa Province of Pakistan. Forty-five isolates of Staphylococcus aureus were characterized by microarray hybridization. Twenty isolates (44 %) were MRSA, whereas 22 (49 %) were PVL-positive. Fourteen isolates (31 %) harboured both mecA and PVL genes. The dominant clones were CC121-MSSA (n = 15, 33 %) and the PVL-positive “Bengal Bay Clone” (ST772-MRSA-V; n = 13, 29 %). The PVL-positive CC8-MRSA-IV strain “USA300” was found once. The pandemic ST239-MRSA-III strain was absent, although it has previously been observed in Pakistan. These observations require a re-assessment of schemes for initial antibiotic therapy to cover MRSA and they emphasise the need for a rapid and non-molecular test for PVL.
Similar content being viewed by others
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a major healthcare problem all over the world. MRSA carries mecA/C genes that confer resistance to beta-lactams on potentially mobile genetic elements named staphylococcal cassette chromosome mec, (SCCmec) [1]. Its high genetic variability, the presence of a variety of different strains [2] and a rapid evolution by acquisition of antibiotic resistance and virulence factors complicate infection control and treatment. A large proportion of recently emerging community-acquired MRSA (CA-MRSA) carries phage-borne genes that encode Panton–Valentine leukocidin (PVL) [3]. This is a bicomponent leucocidin [4] that kills leukocytes, or drives them into apoptosis [5]. It is associated with skin and soft-tissue infections and with necrotising pneumonia [6, 7], warranting more stringent infection control and therapy measures than applied for PVL-negative S. aureus (https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/322857/Guidance_on_the_diagnosis_and_management_of_PVL_associated_SA_infections_in_England_2_Ed.pdf).
Although typing data for Staphylococcus aureus/MRSA are abundantly available for Western Europe, the USA or Australia, comparatively few studies have been published describing the situation in the rest of the world, including Central Asia and Pakistan. Three epidemiological studies confirmed the pandemic healthcare-associated strain ST239-MRSA-III to be prevalent in Pakistan. Zafar et al. [8] analysed 126 samples taken in 2006/2007, mainly from Karachi, using pulsed field gel electrophoresis (PFGE), mecA PCR, SCCmec typing by PCR and multilocus sequence typing (MLST). They found ST8-MRSA-IV and ST239-MRSA-III to be the main genotypes in addition to single isolates of ST1 and ST217 (which belongs to CC22).
In a PhD thesis by Arfat [9] on isolates collected in Rawalpindi and Islamabad between 2006 and 2008, hospital-acquired MRSA (HA-MRSA) was analysed by restriction modification (RM), multiple-locus variable-number tandem repeat analysis (MLVA), SCCmec typing and MLST. Isolates were assigned to ST239, to ST113 (CC8) and to ST30. In another recent study [10], isolates from Rawalpindi and Islamabad were epidemiologically typed by PFGE, staphylococcal interspersed repeat unit typing (SIRU), RM, SCCmec typing, PVL-PCR and MLST. ST239-MRSA-III was found to be the prevalent clone whereas ST8-MRSA-IV, ST113-MRSA-IV and ST30-MRSA-IV [PVL+] were less common.
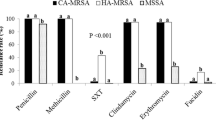
Another study of skin and soft-tissue infections (SSTI) from Abbottabad in 2009/2010 [11] revealed high rates of MRSA, accounting for 28 % of community- and 48 % of hospital-acquired infections. However, no molecular typing was carried out.
Further studies depict CA-MRSA to disseminate in countries such as Iran [12, 13], Kuwait [14, 15], Oman [16], Saudi Arabia [17] and the Gulf Emirates [2, 18]. The amount of PVL-carrying clones is high in these countries as well. Common MRSA clones are ST6-IV, ST1295-IV and ST772-IV in Oman [16]; ST239-III, CC22-IV, CC30-IV and CC80-IV in Saudi Arabia [17]; and ST22-IV, CC8 and CC80-IV in the Gulf Emirates [2, 18]. Recent typing data also exist for India (with ST2371-IV (CC22), ST22-IV, ST772-V and ST8-IV [19–21]) and Iran (where ST239-III, dominated and ST585, ST2732, ST1294, ST30, ST36 and ST1163 were also found [22]).
Here, we report the genotyping of S. aureus, including MSSA, hospital-acquired and community-acquired MRSA, from skin and soft-tissue infections in a (tertiary care) hospital in the Malakand district of the Khyber Pakhtunkhwa Province of Pakistan.
Materials and methods
Sampling area and patient selection
The samples were collected from District Head Quarter Hospital Batkhela Malakand of Khyber Pakhtunkhwa Province between August and September 2015. Study participants were patients in different wards, such as surgical, orthopaedic, dressing room and outpatient departments. Patients having either postsurgical or nosocomial skin and soft-tissue infections or community-acquired skin and soft-tissue infections were selected for sampling. All patients were divided into two broad categories, i.e., those with community-acquired skin and soft-tissue infections (outpatients and patients with infections that manifested within 48 h of admission) and those with healthcare-associated skin and soft-tissue infections (that manifested later than the first 48 h post-admission). All relevant information such as age, gender, occupation, site and type of infection etc. was acquired at the time of sample collection (see also Supplemental Material).
Sampling procedure
The samples were taken using sterile cotton swabs (MWE 170 transwabs; MWE, Corsham, UK). The infection site was first cleansed with alcohol wipes to remove bacteria of the skin’s normal flora. The skin lesion content was absorbed by the transwabs and inserted and sealed in the tube with transport medium. The samples were transported to the microbiology laboratory of the University of Haripur for culturing.
Only one sample per patient was included. If it yielded phenotypically different colonies, both variants were tested separately. They were analysed separately only when genotypically different.
Identification of S. aureus
Mannitol salt agar (MSA; Oxoid, Basingstoke, UK) was used as a selective medium for growing S. aureus. Transwabs with patient samples were streaked on MSA plates and incubated at 37 °C for 24–48 h. S. aureus strains were confirmed by using conventional microbiological tests, i.e. microscopy, catalase and coagulase assays and mannitol fermentation. The S. aureus strains identified were stored in brain heart infusion broth with 20 % glycerol at −20 °C.
Array procedures
The characterization of isolates was performed using StaphyType DNA microarrays (Alere Technologies, Jena, Germany), which cover 333 different target sequences corresponding to approximately 170 distinct genes and their allelic variants. These genes include species markers, typing markers, in addition to toxin genes and resistance genes. Detailed descriptions of genes, sequences and protocols have been published previously [2, 23]. In brief: sample swabs were inoculated on Columbia blood agar. S. aureus was sub-cloned on a Columbia blood agar plate, harvested and enzymatically lysed. DNA was purified using Qiagen spin columns. A linear amplification was then performed with one specific primer for each target. This method allows simultaneous amplification of a multitude of target sequences, but compared with PCR it results in a lower number of copies owing to its linear kinetics. Biotin-dUTP was incorporated into the amplicons during amplification, thus allowing the detection of hybridization to probes immobilized to the array. Detection was performed using streptavidin–horseradish peroxidase, which catalysed a local precipitation of a dye. This resulted in visible spots on the microarray that were imaged and analysed using a designated reader and software (Alere Technologies). The patterns of spots allowed the presence or absence of certain genes or alleles to be established, in addition to, by automated comparison with a database, assignment to clonal complexes, strains and SCCmec types.
Results
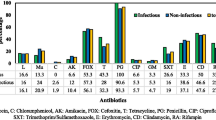
In this study, 45 isolates of S. aureus from skin and soft-tissue infections were characterized. They were assigned to seven clonal complexes and ten distinct strains. Forty-four percent of isolates were MRSA, 49 % were PVL-positive and 31 % harboured both mecA and PVL genes. Tables 1 and 2 show the prevalence data for genes associated with resistance and virulence. Full data are shown in the Supplemental Material.
Assignment to strains and clonal complexes was carried out on the basis of a hybridization pattern, as described previously [2, 23]. Detailed strain descriptions are listed below, and an overview of the distribution of clones and relevant genes is given in Table 3. The dominant clones were CC121-MSSA (33 %) and the PVL-positive “Bengal Bay clone” (ST772-MRSA-V; 29 %). The remaining third of the isolates comprised another eight different strains. ST239 was completely absent.
CC1/ST772-MRSA-V (Bengal Bay clone)
ST772 is usually assigned to CC1 based on the identity of six out of seven MLST genes, but it differs in several key markers such as capsule type (5 in ST772, but 8 in ST1), agr group affiliation (II for ST772, but III for ST1), absence of the enterotoxin gene seh or the presence of the egc enterotoxin gene cluster [24, 25]. Thirteen identical isolates were assigned to this lineage, and to the ST772-MRSA-V strain known as “Bengal Bay clone” [26], all being positive for PVL genes and for SCCmec V. They also harboured enterotoxin genes sea, sec and sel, the gene scn (staphylococcal complement inhibitor) and, for a community-acquired strain, an unusually high number of antibiotic resistance genes, i.e. blaZ (beta-lactamase), msr(A), mph(C) (macrolide resistance), aacA-aphD (gentamicin and tobramycin resistance), aphA3 (kana-/neomycin resistance) and sat (streptothricin resistance). Two out of these 13 isolates were recorded as being hospital-acquired.
CC6-MRSA-IV (WA MRSA-51)
Five isolates were identified as CC6-MRSA-IV, a strain first described in Australia as WA MRSA-51 [27]. All isolates carried sea, but lacked PVL genes, and they did not harbour additional resistance genes beside mecA.
ST8-MRSA-IV [ACME/PVL+] (USA300)
One isolate was assigned to USA300, i.e. PVL- and ACME-positive CC8-MRSA-IV. It appeared to be identical to the fully sequenced strain TCH1516, GenBank CP000730.1.
CC30-MSSA
Seven isolates belonged to CC30-MSSA, 6 of them were PVL-positive. Five of the PVL-positives carried the antibiotic resistance genes blaZ, msr(A), mph(C), aphA3 and sat. A sixth one harboured blaZ, linA (clindamycin/lincosamide resistance), aadD (tobramycin resistance), tetK (tetracycline resistance) in addition to the enterotoxin A gene sea. The PVL-negative isolate lacked all the resistance markers, but also carried sea.
CC121-MSSA
Sixteen isolates belonged to CC121. A single one was PVL-positive, and no CC121-MRSA was detected. All CC121 isolates carried the egc locus and the enterotoxin homologue ORF CM14. The PVL-positive isolate additionally harboured the enterotoxin B gene seb. Fourteen out of 15 PVL-negative isolates carried the genes for exfoliative toxin (etB) and epidermal cell differentiation inhibitor C (edinC). The 15th isolate proved to be a mixed culture of one etB- and edinC-positive variant and a second variant that was negative for these two genes. No resistance genes besides blaZ were detected. A higher rate of hospital-acquired cases was observed for the PVL-negative CC121-MSSA strain than for other strains from this study (Table 3), suggesting a possible in-house transmission.
ST291/813-MSSA
Two isolates showed the characteristic hybridization profile of ST291/813. Although this lineage is—according to MLST—considered a double locus variant of ST398, genome sequencing data indicate that it is separate entity [28, 29]. As CC398, these isolates are agr group I and capsule type 5. However, they do not yield signals for coa and cna, and they differ from ST398 in the alleles of ssl01, ebpS, sdrD and hsdS, and in the presence of leucocidin lukE and spl protease genes. Both isolates harboured blaZ, and genes encoding an exfoliative toxin, etD, and an epidermal cell differentiation inhibitor, edinB; one was PVL-positive.
CC509-MRSA-IV
One isolate of CC509-MRSA-IV was found. It belonged to agr group III and capsule type 8. It carries resistance genes mecA, blaZ, aphA3, sat and tetK. Enterotoxin gene cluster genes selm and selo were detected; other probes for egc genes did not yield signals. Genes ORF CM14 in addition to etB and edinC were present.
Discussion
With regard to MRSA epidemiology, different clones were detected from those in previous studies from Pakistan [8–10]. There, ST239-MRSA-III and ST8-MRSA-IV, in addition to other, less common clones were found.
In our study, we observed a predominance of a PVL-positive ST772-MRSA-V strain. This clone was previously dubbed “Bengal Bay clone” because its first cases were associated with origin from or travel to regions around the Bay of Bengal [26] where this strain became endemic [25, 30, 31], displacing previous predominant HA-MRSA clones in a hospital setting, but also spreading in the community [20]. A local spread has also been reported in Oman [16]. Imported cases were observed in many countries including UK, Ireland, Italy, Norway, Abu Dhabi, the Kingdom of Saudi Arabia, India, New Zealand, Australia, Hong Kong and others [2, 17, 24, 32–35], mostly associated with travel into endemic regions. The distribution of this clone may illustrate an epidemiological link between Pakistan and the Arabian Peninsula, where approximately 2,000,000 Pakistani citizens are employed [36].
Another MRSA strain that was found is CC6-MRSA-IV. This strain was apparently first observed in Australia [27], but also appears to be a common strain in the Middle East. It was found in the Kingdom of Saudi Arabia [17], Kuwait [37], Oman [16] and Abu Dhabi [2, 18] so that a dissemination of epidemic strains in either direction appears to be possible. Interestingly, CC6-MSSA was found to be a common clone in camels in Dubai, thus suggesting a zoonotic transmission from camels to humans and acquisition of a SCCmec element [38].
CC509-MRSA-IV was only once observed in Western Australia, but this isolate originated from a patient without any epidemiological link to Central Asia/Pakistan and differed in carriage of accessory resistance genes (Geoffrey Coombs/Julie Pearson, Perth, WA, Australia; personal communication). Thus, no assumptions can currently be made on the origin and provenance of this strain.
A further sporadic MRSA clone was USA300. This strain is known to be widespread in the USA, Australia and Western Europe [2, 39–43], but for Central Asia, there are no data available yet. Again, a link to the Gulf may be possible, as this strain was detected in Abu Dhabi [2, 18].
However, other MRSA clones that have been reported from the Middle East, such as CC22 or CC80 MRSA, were not found in this study. Most conspicuously, the pandemic ST239-MRSA-III strain was not identified, although earlier studies described it to be common in Pakistan. To confirm its possible demise or disappearance, however, more isolates should be typed and emphasis should be placed on other typical healthcare-associated infections (such as ventilator-associated pneumonia).
With this caveat, our observation may indicate a shift in the population structure of MRSA within the last decade. Although studies from samples taken in 2006 and 2010 revealed ST239, ST8 and ST113 to be the prevalent clones [8–10], these clones are now absent, being replaced largely by the emerging PVL-positive ST772-MRSA-V strain. The ST239 strain used to be a pandemic clone, but it was strictly confined to hospital settings and apparently never spread in the community outside of healthcare facilities. With the emergence of the virulent and rather multi-resistant ST772-MRSA-V strain, MRSA is no longer confined to hospitals, and the abundance of this strain outside of hospitals will make infection control even more difficult. This trend is also observed in other regions [20, 32].
Regarding MSSA, virtually no typing data are available for Pakistan. CC30 and CC121 are both pandemic clones that can essentially be found everywhere [44–51]. A third lineage, ST291/813, was already reported from Pakistan [29], but also from Iran [52], India [53], Mali [54], Romania [51], Algeria and Western Europe [28, 46, 55, 56]. It was also found in dairy milk of Egyptian cows and buffalo [57] and this may indicate a zoonotic origin of this lineage.
The overall results of this study are observations of a high rate of MRSA and a high rate of PVL-positive isolates. There was a high prevalence of both in cases that were classified as community-associated (Table 3). In the wake of these observations a re-assessment of antibiotic therapy schemes are warranted that locally rely on amoxicillin + clavulanic acid, cefixime and ceftriaxone. For instance all patients with ST772-MRSA-V received these beta-lactams (see Supplemental Material). Our observations also emphasize a need for a rapid and non-molecular test for PVL, as described previously [48], and suggest a combined PBP2a + PVL lateral flow assay systems for use in regions with a high burden of MRSA and of PVL-positives, such as Pakistan.
References
Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K (2001) Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 45(5):1323–1336
Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O’Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan H-L, Weber S, Ehricht R (2011) A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6(4), e17936
Vandenesch F, Naimi T, Enright M, Lina G, Nimmo G, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy M (2003) Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 9(8):978–984
Kaneko J, Kamio Y (2004) Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci Biotechnol Biochem 68(5):981–1003
Genestier AL, Michallet MC, Prevost G, Bellot G, Chalabreysse L, Peyrol S, Thivolet F, Etienne J, Lina G, Vallette FM, Vandenesch F, Genestier L (2005) Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J Clin Invest 115(11):3117–3127
Francis JS, Doherty MC, Lopatin U, Johnston CP, Sinha G, Ross T, Cai M, Hansel NN, Perl T, Ticehurst JR, Carroll K, Thomas DL, Nuermberger E, Bartlett JG (2005) Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis 40(1):100–107
Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Hook M, Etienne J, Vandenesch F, Bowden MG (2007) Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science 315(5815):1130–1133
Zafar A, Stone M, Ibrahim S, Parveen Z, Hasan Z, Khan E, Hasan R, Wain J, Bamford K (2011) Prevalent genotypes of meticillin-resistant Staphylococcus aureus: report from Pakistan. J Med Microbiol 60(Pt 1):56–62
Arfat Y (2013) Genotyping of methicillin resistant Staphylococcus aureus (MRSA) from local hospital of Rawalpindi/Islamabad, Pakistan. PhD, Quaid-i-Azam University, Islamabad
Shabir S, Hardy KJ, Abbasi WS, McMurray CL, Malik SA, Wattal C, Hawkey PM (2010) Epidemiological typing of meticillin-resistant Staphylococcus aureus isolates from Pakistan and India. J Med Microbiol 59(Pt 3):330–337. doi:10.1099/jmm.0.014910-0
Ahmad MK, Asrar A (2014) Prevalence of methicillin resistant Staphylococcus aureus in pyogenic community and hospital acquired skin and soft tissues infections. J Pak Med Assoc 64(8):892–895
Mamishi S, Mahmoudi S, Bahador A, Matini H, Movahedi Z, Sadeghi RH, Pourakbari B (2015) Emergence of community-acquired methicillin-resistant Staphylococcus aureus in an Iranian referral paediatric hospital. Br J Biomed Sci 72(2):47–51
Havaei S, Moghadam SO, Pourmand M, Faghri J (2010) Prevalence of genes encoding bi-component leukocidins among clinical isolates of methicillin resistant Staphylococcus aureus. Iran J Public Health 39(1):8–14
Udo E, O’Brien F, Al-Sweih N, Noronha B, Matthew B, Grubb W (2008) Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J Clin Microbiol 46(10):3514–3516
Udo EE, Al-Sweih N, Noronha B (2006) Characterisation of non-multiresistant methicillin-resistant Staphylococcus aureus (including EMRSA-15) in Kuwait Hospitals. Clin Microbiol Infect 12(3):262–269
Udo EE, Al-Lawati BAH, Al-Muharmi Z, Thukral SS (2014) Genotyping of methicillin-resistant Staphylococcus aureus in the Sultan Qaboos University Hospital, Oman reveals the dominance of Panton–Valentine leucocidin-negative ST6-IV/t304 clone. New Microbes New Infect 2(4):100–105. doi:10.1002/nmi2.47
Monecke S, Skakni L, Hasan R, Ruppelt A, Ghazal SS, Hakawi A, Slickers P, Ehricht R (2012) Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol 12(1):146
Weber S, Ehricht R, Slickers P, Abdel-Wareth L, Donnelly G, Pitout M, Monecke S (2010) Genetic fingerprinting of MRSA from Abu Dhabi. In: ECCMID: 2010; Vienna
Rajan V, Schoenfelder SMK, Ziebuhr W, Gopal S (2015) Genotyping of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) in a tertiary care centre in Mysore, South India: ST2371-SCCmec IV emerges as the major clone. Infect Genet Evol 34:230–235. doi:10.1016/j.meegid.2015.05.032
D’Souza N, Rodrigues C, Mehta A (2010) Molecular characterization of methicillin resistant Staphylococcus aureus (MRSA) with emergence of epidemic clones ST 22 and ST 772, in Mumbai, India. J Clin Microbiol 48(5):1806–1811
Nadig S, Sowjanya S, Seetharam S, Bharathi K, Raghunath D, Arakere G (2009) Molecular characterization of Indian methicillin resistant Staphylococcus aureus. Proceedings of the Ninth Sir Dorabji Tata Symposium on Antimicrobial resistance—the modern epidemic: Current Status and Research Issues: 10–11 March 2008, pp 167–184
Ohadian Moghadam S, Pourmand MR, Mahmoudi M, Sadighian H (2015) Molecular characterization of methicillin-resistant Staphylococcus aureus: characterization of major clones and emergence of epidemic clones of sequence type (ST) 36 and ST 121 in Tehran, Iran. FEMS Microbiol Lett 362(8):fnv043. doi:10.1093/femsle/fnv043
Monecke S, Slickers P, Ehricht R (2008) Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol 53:237–251
Monecke S, Baier V, Coombs GW, Slickers P, Ziegler A, Ehricht R (2013) Genome sequencing and molecular characterisation of Staphylococcus aureus ST772-MRSA-V, “Bengal Bay Clone”. BMC Res Notes 6(1):1–7. doi:10.1186/1756-0500-6-548
Prabhakara S, Khedkar S, Loganathan RM, Chandana S, Gowda M, Arakere G, Seshasayee ASN (2012) Draft genome sequence of Staphylococcus aureus 118 (ST772), a major disease clone from India. J Bacteriol 194:3727–3728. doi:10.1128/jb.00480-12
Ellington MJ, Ganner M, Warner M, Cookson BD, Kearns AM (2010) Polyclonal multiply antibiotic-resistant methicillin-resistant Staphylococcus aureus with Panton-Valentine leucocidin in England. J Antimicrob Chemother 65(1):46–50
Coombs G, Pearson J, Daley D, Robinson O, Nimmo G, Turnidge JD (2013) Staphylococcus aureus Programme 2012 (SAP 2012) Community Survey, MRSA Epidemiology and Typing Report. Perth, Gold Coast, Adelaide
Stegger M, Aziz M, Chroboczek T, Price LB, Ronco T, Kiil K, Skov RL, Laurent F, Andersen PS (2013) Genome analysis of Staphylococcus aureus ST291, a double locus variant of ST398, reveals a distinct genetic lineage. PLoS One 8(5), e63008. doi:10.1371/journal.pone.0063008
Marasa BS, Khan S, Iram S, Sung K, Xu J (2015) Draft genome sequence of methicillin-resistant clinical Staphylococcus aureus isolate 51S (sequence type 291). Genome Announc 3(5):e01050-15. doi:10.1128/genomeA.01050-15
Afroz S, Kobayashi N, Nagashima S, Alam MM, Hossain AB, Rahman MA, Islam MR, Lutfor AB, Muazzam N, Khan MA, Paul SK, Shamsuzzaman AK, Mahmud MC, Musa AK, Hossain MA (2008) Genetic characterization of Staphylococcus aureus isolates carrying Panton-Valentine leukocidin genes in Bangladesh. Jpn J Infect Dis 61(5):393–396
Prabhakara S, Khedkar S, Shambat SM, Srinivasan R, Basu A, Norrby-Teglund A, Seshasayee ASN, Arakere G (2013) Genome sequencing unveils a novel sea enterotoxin-carrying PVL phage in Staphylococcus aureus ST772 from India. PLoS One 8(3):e60013. doi:10.1371/journal.pone.0060013
Brennan GI, Shore AC, Corcoran S, Tecklenborg S, Coleman DC, O’Connell B (2012) Emergence of hospital- and community-associated Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus genotype ST772-MRSA-V in Ireland and detailed investigation of an ST772-MRSA-V cluster in a neonatal intensive care unit. J Clin Microbiol 50(3):841–847. doi:10.1128/jcm.06354-11
Sanchini A, Campanile F, Monaco M, Cafiso V, Rasigade JP, Laurent F, Etienne J, Stefani S, Pantosti A (2011) DNA microarray-based characterisation of Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus from Italy. Eur J Clin Microbiol Infect Dis 30(11):1399–1408. doi:10.1007/s10096-011-1234-x
Williamson DA, Roberts SA, Ritchie SR, Coombs GW, Fraser JD, Heffernan H (2013) Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in New Zealand: rapid emergence of sequence type 5 (ST5)-SCCmecIV as the dominant community-associated MRSA clone. PLoS One 8(4), e62020. doi:10.1371/journal.pone.0062020
Monecke S, Aamot HV, Stieber B, Ruppelt A, Ehricht R (2014) Characterization of PVL-positive MRSA from Norway. APMIS 122(7):580–584. doi:10.1111/apm.12181
Government of Pakistan MoL, Manpower & Overseas Pakistanis (2005) Year Book 2004–2005
Udo EE, Al-Sweih N (2013) Emergence of methicillin-resistant Staphylococcus aureus in the maternity hospital, Kuwait. Med Princ Pract 22(6):535–539
Monecke S, Ehricht R, Slickers P, Wernery R, Johnson B, Jose S, Wernery U (2011) Microarray-based genotyping of Staphylococcus aureus isolates from camels. Vet Microbiol 150(3–4):309–314
Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367(9512):731–739
Kourbatova EV, Halvosa JS, King MD, Ray SM, White N, Blumberg HM (2005) Emergence of community-associated methicillin-resistant Staphylococcus aureus USA 300 clone as a cause of health care-associated infections among patients with prosthetic joint infections. Am J Infect Control 33(7):385–391
Larsen A, Stegger M, Goering R, Sorum M, Skov R (2007) Emergence and dissemination of the methicillin resistant Staphylococcus aureus USA300 clone in Denmark (2000–2005). Euro Surveill 12(2)
Monecke S, Ehricht R, Slickers P, Tan HL, Coombs G (2009) The molecular epidemiology and evolution of the Panton-Valentine leukocidin-positive, methicillin-resistant Staphylococcus aureus strain USA300 in Western Australia. Clin Microbiol Infect 15(8):770–776
Otter JA, Havill NL, Boyce JM, French GL (2009) Comparison of community-associated methicillin-resistant Staphylococcus aureus from teaching hospitals in London and the USA, 2004–2006: where is USA300 in the UK? Eur J Clin Microbiol Infect Dis 28(7):835–839
Conceicao T, Santos Silva I, de Lencastre H, Aires-de-Sousa M (2014) Staphylococcus aureus nasal carriage among patients and health care workers in Sao Tome and Principe. Microb Drug Resist 20(1):57–66. doi:10.1089/mdr.2013.0136
Egyir B, Guardabassi L, Sorum M, Nielsen SS, Kolekang A, Frimpong E, Addo KK, Newman MJ, Larsen AR (2014) Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from healthcare institutions in Ghana. PLoS One 9(2), e89716. doi:10.1371/journal.pone.0089716
Rasigade J-P, Laurent F, Lina G, Meugnier H, Bes M, Vandenesch F, Etienne J, Tristan A (2010) Global distribution and evolution of Panton-Valentine leukocidin-positive methicillin-susceptible Staphylococcus aureus, 1981–2007. J Infect Dis 201(10):1589–1597. doi:10.1086/652008
Vorobieva V, Bazhukova T, Hanssen AM, Caugant DA, Semenova N, Haldorsen BC, Simonsen GS, Sundsfjord A (2008) Clinical isolates of Staphylococcus aureus from the Arkhangelsk region, Russia: antimicrobial susceptibility, molecular epidemiology, and distribution of Panton-Valentine leukocidin genes. APMIS 116(10):877–887
Monecke S, Muller E, Buechler J, Rejman J, Stieber B, Akpaka PE, Bandt D, Burris R, Coombs G, Hidalgo-Arroyo GA, Hughes P, Kearns A, Abos SM, Pichon B, Skakni L, Soderquist B, Ehricht R (2013) Rapid detection of Panton-Valentine leukocidin in Staphylococcus aureus cultures by use of a lateral flow assay based on monoclonal antibodies. J Clin Microbiol 51(2):487–495. doi:10.1128/JCM.02285-12
Monecke S, Slickers P, Ellington M, Kearns A, Ehricht R (2007) High diversity of Panton-Valentine leucocidin-positive, methicillin-susceptible isolates of Staphylococcus aureus and implications for the evolution of community-associated MRSA. Clin Microbiol Infect 13(12):1157–1164
Robinson DA, Kearns AM, Holmes A, Morrison D, Grundmann H, Edwards G, O’Brien FG, Tenover FC, McDougal LK, Monk AB, Enright MC (2005) Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet 365(9466):1256–1258
Monecke S, Müller E, Dorneanu OS, Vremeră T, Ehricht R (2014) Molecular typing of MRSA and of clinical Staphylococcus aureus isolates from Iaşi Romania. PLoS One. doi:10.1371/journal.pone.0097833
Havaei SA, Azimian A, Fazeli H, Naderi M, Ghazvini K, Samiee SM, Soleimani M (2013) Isolation of Asian endemic and livestock associated clones of methicillin resistant Staphylococcus aureus from ocular samples in Northeastern Iran. Iran J Microbiol 5(3):227–232
Shambat S, Nadig S, Prabhakara S, Bes M, Etienne J, Arakere G (2012) Clonal complexes and virulence factors of Staphylococcus aureus from several cities in India. BMC Microbiol 12:64. doi:10.1186/1471-2180-12-64
Ruimy R, Maiga A, Armand-Lefevre L, Maiga I, Diallo A, Koumare AK, Ouattara K, Soumare S, Gaillard K, Lucet JC, Andremont A, Feil EJ (2008) The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J Bacteriol 190(11):3962–3968
Sakwinska O, Kuhn G, Balmelli C, Francioli P, Giddey M, Perreten V, Riesen A, Zysset F, Blanc DS, Moreillon P (2009) Genetic diversity and ecological success of Staphylococcus aureus strains colonizing humans. Appl Environ Microbiol 75(1):175–183
Green SM, Marsh P, Ahmad N, Jefferies JMC, Clarke SC (2010) Characterization of community and hospital Staphylococcus aureus isolates in Southampton, UK. J Med Microbiol 59(9):1084–1088. doi:10.1099/jmm.0.018986
El-Ashker M, Gwida M, Tomaso H, Monecke S, Ehricht R, El-Gohary F, Hotzel H (2015) Staphylococci in cattle and buffaloes with mastitis in Dakahlia Governorate, Egypt. J Dairy Sci 98(11):7450–7459. doi:10.3168/jds.2015-9432
Acknowledgements
The authors thank the Vice Chancellor’s Office of the University of Haripur (Pakistan) for supporting this work. We also acknowledge Ines Engelmann (Alere Technologies, Jena, Germany) for technical help.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Sara Madzgalla, Elke Müller, Annett Reissig, Ralf Ehricht and Stefan Monecke are employees of Alere Technologies, Jena, Germany, the company that manufactures the microarrays used for this study.
Funding information
There was no external funding for this work. Each of the participating institutions covered the costs of experiments performed in the respective institutions, and granted the time needed to perform this study.
Informed consent and ethical approval
Informed written consent was taken from each patient and the study was approved by the institutional ethics committee.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 235 kb)
Rights and permissions
About this article
Cite this article
Madzgalla, S., Syed, M.A., Khan, M.A. et al. Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections in patients from Malakand, Pakistan. Eur J Clin Microbiol Infect Dis 35, 1541–1547 (2016). https://doi.org/10.1007/s10096-016-2695-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2695-8