Abstract
The purpose of this investigation was the determination of the distribution of genotypes and alleles, residing within interleukin 6 (IL6) and interleukin 1 (IL1) polymorphisms, among fetuses and neonates, congenitally infected with Toxoplasma gondii, and among uninfected control cases. The study included 22 fetuses and newborns infected with T. gondii and 49 control cases. Screening for IgG and IgM antibodies against the parasite and IgG avidity was performed by enzyme-linked fluorescent assay (ELFA) tests. Quantitation of T. gondii DNA in amniotic fluids was assayed by the real-time Q PCR technique for the parasitic B1 gene. Genotypes at IL6 and IL1 single nucleotide polymorphisms (SNPs) were determined by a self-designed, nested polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay. Representative genotypes at the studied loci were confirmed by sequencing. All the genotypes were estimated for Hardy–Weinberg equilibrium and IL1 genotypes were tested for linkage disequilibrium. Genotypes and haplotypes at the studied SNPs were investigated for their possible association with the occurrence of congenital T. gondii infection, using a logistic regression model. GC heterozygotes at the IL6 −174 G>C SNP were significantly associated with toxoplasmosis and increased the risk of T. gondii infection [odds ratio (OR) 4.24, 95 % confidence interval (CI) 1.24–14.50 in the codominant model, p ≤ 0.050]. In case of IL1 SNPs, similar prevalence rates were observed between T. gondii-infected and -uninfected offspring. Regarding allelic variability, the C alleles at both IL6 and IL1B SNPs were significantly more frequent in the infected than in the uninfected cases (p ≤ 0.050). It is concluded that IL6 −174 G>C and IL1B +3954 C>T SNPs might be involved in the development of congenital T. gondii infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Toxoplasma gondii is one of the most common intrauterine infections worldwide and the major cause of perinatal morbidity and mortality [1–4]. Congenital infections with T. gondii are diagnosed in about 0.07–2.9 % of live newborns, leading to asymptomatic as well as symptomatic disease, sometimes with fatal course [1, 5, 6].
Considering the immune response against T. gondii, several studies have shown the role of proinflammatory cytokines, including interleukin (IL) 6 and IL1 [7–9]. Among women infected with T. gondii, IL6 expression was estimated to be twice as high compared to the control cases [10]. In another study, the development of toxoplasmosis-related ocular lesions among T. gondii-infected patients was reported to be associated with high IL1 and TNF-α levels [11]. In turn, the mice with more severe inflammation in the retina and vitreous humor, related to ocular toxoplasmosis, had a reduced expression of ocular IL1A and an increased production of TNFA mRNA as compared to WT mice [12]. In an in vitro study, the human monocytic wild-type MonoMac6 cells, infected with T. gondii, showed significantly higher levels of IL-1β as compared to non-infected cells [13]. Considering a possible participation of the genetic alterations, located at IL1 and IL6 molecule encoding genes, the −174 G>C single nucleotide polymorphism (SNP) from the IL6 gene was reported to be correlated with toxoplasmic retinochoroiditis (TR) [14]. The prevalence rates of genotypes and alleles at the IL6 −174 G>C SNP differed significantly between the patients with TR and the healthy blood donors with positive serology for T. gondii infection and without retinal symptoms of the previous disease [14]. In turn, another study reported no correlation of either IL1A −889 C>T or IL1B C>T SNPs with the occurrence of TR [15]. However, distinct differences in the distribution of genotypes and alleles at the IL1A SNP were observed between patients with and without recurrent episodes of the disease [15]. So far, no study has been performed to seek for a possible involvement of IL1 and IL6 SNPs in the development of congenital infection with T. gondii.
In the reported study, we aimed to analyze and describe a possible influence of the polymorphisms located at the IL1A, IL1B, and IL6 genes on the occurrence of congenital T. gondii infection in fetuses and neonates. The haplotype prevalence rates were also estimated for IL1 SNPs. A multiple-SNP analysis was performed to estimate the assumed simultaneous influence of IL1 and IL6 SNPs on the occurrence of the disease.
Materials and methods
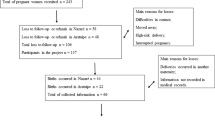
The study included 22 fetuses congenitally infected with T. gondii and 49 uninfected control cases, from which samples were retrospectively (16 infected cases and 25 controls) as well as prospectively (six infected cases and 24 controls) collected. The fetal and neonatal specimens were obtained from the offspring of pregnant women treated at the Department of Fetal-Maternal Medicine and Gynecology at the Polish Mother’s Memorial Hospital—Research Institute in Lodz, between the years 2000 and 2014. Clinical samples, selected for genetic studies, consisted of fetal amniotic fluids, obtained via amniocentesis from pregnant women, and/or cerebrospinal fluids (six samples), as well as umbilical cord blood specimens (two samples). The intrauterine infections with T. gondii were determined from maternal serological features of the recent infection and by the clinical picture, including flu-like symptoms, as well as by fetal and neonatal ultrasound markers associated with toxoplasmosis. The congenital infections with T. gondii were confirmed by the presence of parasitic DNA in fetal and neonatal body fluids. The research was approved by the Research Ethics Committee at the Polish Mother’s Memorial Hospital—Research Institute. All the samples, previously collected for diagnostic purposes, are anonymized in this report. Informed consent forms were signed by all the enrolled participants (pregnant women).
Serological tests
Blood specimens were collected from the pregnant women by venipuncture on their first visit to the hospital. Serum samples were obtained by centrifugation and then stored at 4 °C until analysis. Serological tests were performed at the hospital’s Department of Clinical Microbiology.
Screening for T. gondii IgG antibodies was performed with the enzyme-linked fluorescent assay (ELFA) VIDAS TOXO IgG II (bioMérieux). The IgM antibodies were detected with the ELFA assay VIDAS TOXO IgM (bioMérieux). T. gondii IgG avidity was assayed by an ELFA assay VIDAS TOXO IgG AVIDITY (bioMérieux). The pregnant women were diagnosed as recently infected with T. gondii in case of seroconversion during pregnancy or were suspected to be infected in case of the infection-related serology including IgM seropositivity and low IgG avidity index. Fresh intrauterine T. gondii infections, possible in the studied pregnant women, were confirmed by real-time Q PCR assays for the parasitic B1 gene fragments, performed on body fluid specimens of their offspring.
DNA extraction
Fetal genomic DNA was extracted from 5-mL amniotic fluid volumes or 3-mL cerebrospinal fluid volumes, and neonatal DNA was extracted from 200 μL of umbilical cord blood specimens, using a High Pure PCR Template Preparation Kit (Roche™, Mannheim, Germany). The obtained DNA was diluted in 100 μL of elution buffer and stored at −20 °C until further molecular analyses.
Detection and quantification of T. gondii loads
The amount of T. gondii DNA was quantified by a real-time (RT) Q PCR assay for the detection of parasitic B1 gene fragment of 83 bps in length. Forward and reverse primers and TaqMan MGB probe sequences were as follows: 5′-CAAGCAGCGTATTGTCGAGTAGAT-3′, 5′-GCGTCTCTTTCATTCCCACATTTT-3′, and 5′-6-FAM- CAGAAAGGAACTGCATCCGTT-NFQ-3′, respectively [16]. The reactions were prepared in final 25-μL volumes, as described previously [17].
Determination of SNPs located within IL1 and IL6 genes
IL1A −889 C>T, IL1B +3954 C>T, and IL6 −174 G>C SNPs were genotyped, using self-designed nested polymerase chain reaction (PCR) assays. The used external and internal primer sequences, amplicon lengths, and annealing temperatures are shown in Table 1. External primer sequences were developed using the Vector NTI Suite 5.5 software, while the internal primers were based on published data [14, 18–21]. The amplifications were performed with a HotStarTaq® Master Mix Kit (Qiagen, Hilden, Germany). The PCR conditions were as follows: an initial activation for 15 min at 95 °C and for 40 cycles of repeated denaturation at 94 °C for 30 s, annealing at temperatures designed for the used primer pairs (see Table 1), for 1 min, and extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. The nested PCR products were resolved on 1 % agarose gels, stained with ethidium bromide, and then digested overnight with NcoI, TaqI, and Hsp92II endonucleases, used to determine genotypes at IL1A −889 C>T, IL1B +3954 C>T, and IL6 −174 G>C SNPs, respectively (see Table 2). The products of digestion reactions were resolved on 2 % agarose gels. Genotypes were determined at the analyzed polymorphic sites by the length of restriction fragments, as described previously ([14, 18–21]; see Table 2, Fig. 1). The randomly selected PCR products, including the analyzed SNPs, were then verified by sequencing according to the Sanger method. In case of IL1A and IL1B SNPs, the sequencing was performed for seven and nine CC homozygotes, four and five CT heterozygotes, as well as for three and one TT homozygotes, respectively. Regarding the IL6 SNP, sequencing was done for five GG homozygotes, five GC heterozygotes, and 12 CC homozygotes. Examples of chromatograms with DNA sequences of distinct genotypes at the studied polymorphisms are shown in Figs. 2 and 3. The sequenced and referenced DNA regions were compared using the BLASTN program for alignment of two (or more) sequences. The chromatograms were read out using the Sequence Scanner 1.0 and Chromas Lite 2.1.1 softwares.
Agarose gel electrophoresis of polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) products for distinct genotypes at IL1A −889 C>T single nucleotide polymorphism (SNP) (a), IL1B +3954 C>T SNP (b), and IL6 −174 G>C SNP (c). DNA fragments, obtained by the restriction analyses with NcoI (a), TaqI (b), and Hsp92II (c) endonucleases, were resolved in 2 % agarose gels, stained with ethidium bromide. The numbers on the right indicate the size of separated DNA fragments. M 50-bp DNA marker; Ud undigested PCR product; CC, CT, TT, GG, GC, CC genotypes at the studied IL polymorphisms
Statistical analysis
The prevalence rates of genotypes and alleles at the IL1 and IL6 polymorphisms in the infected and uninfected fetuses and neonates were estimated by means of descriptive statistics. The analyzed groups of patients were studied for the Hardy–Weinberg (H-W) equilibrium, the linkage disequilibrium (LD), and haplotypes, using the SNPStats software (http://bioinfo.iconcologia.net/en/SNPStats_web). The correlation of the genotypes, alleles, or haplotypes at IL SNPs with the development of congenital T. gondii infections was determined by cross-tabulation, Pearson’s Chi-squared, as well as by the logistic regression model. The analysis of haplotypes at IL1 SNPs and the multiple-SNP analysis were carried out by the expectation–maximization (EM) algorithm. The study results were estimated as being statistically significant when the significance level was p ≤ 0.050. The statistical analysis was, in part, supported by the NCSS 97 software.
Results
Hardy–Weinberg equilibrium, linkage disequilibrium
The genotypes at all the analyzed polymorphic sites preserved the H-W equilibrium in the fetuses and neonates infected with T. gondii (p = 0.081, p = 0.430, and p = 0.680, for IL1A −889 C>T, IL1B +3954 C>T, and IL6 −174 G>C SNPs, respectively). Taking into account the control cases, the genotypes at IL1B +3954 C>T were in H-W equilibrium (p = 0.150), while the variants at IL1A −889 C>T and IL6 −174 G>C were not (p ≤ 0.050). IL1A −889 C>T and IL1B +3954 C>T SNPs were seen in LD between the infected and the control groups of patients (p ≤ 0.050).
Prevalence rates of the genotypes at IL1A, IL1B, and IL6 SNPs
In the offspring infected with T. gondii, the prevalence rates of CC, CT, and TT genotypes at IL1A −889 C>T/IL1B C>T SNPs were 63.6 % (14/22)/72.7 % (16/22), 22.7 % (5/22)/22.7 % (5/22), and 13.6 % (3/22)/4.5 % (1/22), respectively (see Table 3). Among the control cases, the prevalence rates of the analyzed variants were 51.0 % (25/49)/57.1 % (28/49), 26.5 % (13/49)/30.6 % (15/49), and 22.4 % (11/49)/12.2 % (6/49), respectively. Taking into account the IL6 −174 G>C SNP, GG, GC, and CC genotypes were carried by 27.3 % (6/22), 45.5 % (10/22), and 27.3 % (6/22) of the infected offspring, respectively. In control cases, the prevalence rates of GG, GC, and CC variants at the IL6 SNP were 57.1 % (28/49), 22.4 % (11/49), and 20.4 % (10/49), respectively. Considering the IL6 SNP, the GC heterozygotes were significantly associated with toxoplasmosis and increased the risk of T. gondii infection [odds ratio (OR) 4.24, 95 % confidence interval (CI) 1.24–14.50 in the codominant model, and OR 3.56, 95 % CI 1.19–10.64 for GC and CC genotypes in the dominant model; p ≤ 0.050; see Table 3]. Regarding the IL1A and IL1B SNPs, the observed genotypes were distributed similarly between the infected and uninfected offspring. Neither the haplotype analysis performed for IL1 SNPs nor the multiple-SNP analysis for all the analyzed polymorphisms showed any differences in the prevalence rates of distinct variants between the studied groups.
Distribution of the alleles at IL1 and IL6 SNPs
Among the infected fetuses and neonates, the prevalence rates of C and T alleles at IL1A/IL1B SNPs were 75.0 % (33/44)/84.1 % (37/44) and 25.0 % (11/44)/15.9 % (7/44), respectively (see Table 4). In case of the uninfected patients, the analyzed alleles were found in 64.3 % (63/98)/72.4 % (71/98) and 35.7 % (35/98)/27.6 % (27/98) of the offspring, respectively. Taking into account the IL6 SNP, the G and C alleles were identified in 50 % (22/44) of the infected cases for each allele. In the uninfected controls, the prevalence rates of G and C alleles were 68.4 % (67/98) and 31.6 % (31/98), respectively. A cross-tabulation analysis showed that alleles at the IL1A polymorphic site were similarly distributed between T. gondii-infected and -uninfected fetuses and neonates (see Table 4). In case of the IL1B SNP, the major C allele was significantly more frequent among the infected than among the uninfected offspring (χ2 = 7, p ≤ 0.050; Fisher’s exact test). Considering the IL6 SNP, the minor C allele was significantly more frequent among the infected versus the uninfected cases (χ2 = 4, p ≤ 0.050; Fisher’s exact test).
Discussion
In our study, we found that infants and neonates with GC genotypes at the IL6 −174 G>C SNP were at an approximately four times higher risk of congenital infection with T. gondii as compared to GG and CC homozygotes. Moreover, the minor C allele at the studied locus was significantly more prevalent among the infected than in the uninfected offspring. Regarding T. gondii infections, only one previous study, performed for patients with TR and control cases, reported the role of the IL6 −174 G>C polymorphism for the occurrence of disease [14]. Similarly to our results, the prevalence rates of distinct genotypes at the analyzed IL6 locus were significantly different between the study groups of patients and control cases. In patients with TR, the GC heterozygotic status at the SNP was estimated to be correlated to occurrence of the disease as well [14]. The significantly higher prevalence rate of the C allele versus the G allele observed in our study among the T. gondii-infected offspring as compared to the control cases was also estimated for TR patients [14]. Previously, the IL6 −174 G>C polymorphism was determined as being associated with altered expression levels of the encoded cytokine as well [22–24]. Studies on the role of the IL6 −174 G>C SNP in RSV-infected macrophages showed GC heterozygotes and CC homozygotes as high IL6 production genotypes, correlated with the infection, while among RSV-infected patients, the CC genotype at the analyzed SNP was associated with low IL6 levels [22, 23]. Taking into account the papers reporting increased IL6 levels to be correlated with T. gondii infection in various patient groups, it seems possible that the GC heterozygotic status at the IL6 −174 G>C SNP may alter the immune response against the parasite through higher IL6 cytokine production. However, a detailed description of IL6 molecular changes, possibly associated with congenital toxoplasmosis development, would be a challenge. So far, the C allele at the IL6 −174G>C SNP was reported to generate new binding sites for NF-1 and Smad4 transcription factors, which are not observed in the presence of the G allele [25, 26]. In addition, a variable AnTn tract, located at the nucleotides from −392 up to −373 within the IL6 gene promoter, was also reported to affect the transcription of −174 G allele and −174 C allele-related haplotypes [27].
According to genetic modifications localized in IL1 genes, we found the major C allele at the IL1B +3954 C>T SNP to be significantly more frequent among the fetuses and neonates with congenital T. gondii infection than among the uninfected cases. So far, several studies showed some involvement of the proinflammatory IL1β cytokine in an immune response after T. gondii infection, although the results are ambiguous [8, 11, 28]. Taking into account IL1 polymorphisms, a previous study in TR patients showed, likewise to our outcomes, similar distributions of the genotypes and, in addition, of the alleles at the analyzed IL1A −889 C>T and IL1B +3954 C>T SNPs in patients with TR and in control cases [15]. The outcomes obtained in our study may suggest that the presence of mutated T allele at the analyzed IL1B SNP plays a protective function against the development of congenital toxoplasmosis. However, the mechanism of the role of the IL1B 3954 C>T SNP should be investigated in a detailed molecular study.
Considering our outcomes, as presented in this paper, the GC heterozygotic status at the IL6 −174 G>C SNP, as well as the presence of C alleles at both IL6 and IL1B +3954 C>T SNPs, seems to have been involved in the occurrence of congenital toxoplasmosis in the studied fetuses and neonates. So far, several studies have shown some contribution of the encoded IL6 and IL1β cytokines to immune responses against T. gondii, although no such research has been performed for congenital toxoplasmosis. In case of IL6, the GC heterozygotic status at the −174 G>C SNP was determined to be correlated with TR development, while no such relationship was observed for either the IL1A −889 C>T or IL1B +3954 C>T SNPs. Based on our outcomes, we suggest a possible contribution of GC heterozygotic status at the IL6 −174 G>C SNP to the immune response efficacy against congenital T. gondii infection via an increased synthesis of the IL6 cytokine, whereas the T allele at the IL1B +3954 C>T SNP might be protective against the infection. Although the character of observed genetic changes is suggestive of their effective relationship with congenital toxoplasmosis, a detailed further mechanistic research should be undertaken to investigate the hypothesis in detail. We also suggest that further studies on the role of IL6 and IL1 SNPs, performed with larger groups of patients with congenital toxoplasmosis, would be beneficial to verify our results.
References
Berrebi A, Bardou M, Bessieres MH, Nowakowska D, Castagno R, Rolland M, Wallon M, Franck J, Bongain A, Monnier-Barbarino P, Assouline C, Cassaing S (2007) Outcome for children infected with congenital toxoplasmosis in the first trimester and with normal ultrasound findings: a study of 36 cases. Eur J Obstet Gynecol Reprod Biol 135(1):53–57
Nowakowska D, Colón I, Remington JS, Grigg M, Gołąb E, Wilczyński J, Sibley LD (2006) Genotyping of Toxoplasma gondii by multiplex PCR and peptide-based serological testing of samples from infants in Poland diagnosed with congenital toxoplasmosis. J Clin Microbiol 44(4):1382–1389
Nowakowska D, Wujcicka W, Sobala W, Śpiewak E, Gaj Z, Wilczyński J (2014) Age-associated prevalence of Toxoplasma gondii in 8281 pregnant women in Poland between 2004 and 2012. Epidemiol Infect 142(3):656–661
Wujcicka W, Wilczyński J, Nowakowska D (2013) SNPs in toll-like receptor (TLR) genes as new genetic alterations associated with congenital toxoplasmosis? Eur J Clin Microbiol Infect Dis 32(4):503–511
Remington JS, McLeod R, Wilson CB, Desmonts G (2011) Toxoplasmosis. In: Remington JS, Klein JO, Wilson CB, Nizet V, Maldonado YA (eds) Infectious diseases of the fetus and newborn infant, 7th edn. Elsevier Saunders, Philadelphia, pp 918–1041
Nowakowska D, Respondek-Liberska M, Gołąb E, Stray-Pedersen B, Szaflik K, Dzbenski TH, Wilczyński J (2005) Too late prenatal diagnosis of fetal toxoplasmosis: a case report. Fetal Diagn Ther 20(3):190–193
Jebbari H, Roberts CW, Ferguson DJ, Bluethmann H, Alexander J (1998) A protective role for IL-6 during early infection with Toxoplasma gondii. Parasite Immunol 20(5):231–239
Marshall ES, Elshekiha HM, Hakimi MA, Flynn RJ (2011) Toxoplasma gondii peroxiredoxin promotes altered macrophage function, caspase-1-dependent IL-1beta secretion enhances parasite replication. Vet Res 42:80
Prada J, Liekfeld A, Bergmann F, Grobusch MP (2007) Expression of interleukin-6, tumour necrosis factor-alpha and nitric oxides during episodes of ocular toxoplasmosis in an HIV patient. Acta Ophthalmol Scand 85(8):911–913
Matowicka-Karna J, Dymicka-Piekarska V, Kemona H (2009) Does Toxoplasma gondii infection affect the levels of IgE and cytokines (IL-5, IL-6, IL-10, IL-12, and TNF-alpha)? Clin Dev Immunol 2009:374696
Yamamoto JH, Vallochi AL, Silveira C, Filho JK, Nussenblatt RB, Cunha-Neto E, Gazzinelli RT, Belfort R Jr, Rizzo LV (2000) Discrimination between patients with acquired toxoplasmosis and congenital toxoplasmosis on the basis of the immune response to parasite antigens. J Infect Dis 181(6):2018–2022
Lyons RE, Anthony JP, Ferguson DJ, Byrne N, Alexander J, Roberts F, Roberts CW (2001) Immunological studies of chronic ocular toxoplasmosis: up-regulation of major histocompatibility complex class I and transforming growth factor beta and a protective role for interleukin-6. Infect Immun 69(4):2589–2595
Witola WH, Mui E, Hargrave A, Liu S, Hypolite M, Montpetit A, Cavailles P, Bisanz C, Cesbron-Delauw MF, Fournié GJ, McLeod R (2011) NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect Immun 79(2):756–766
Cordeiro CA, Moreira PR, Bessa TF, Costa GC, Dutra WO, Campos WR, Oréfice F, Young LH, Teixeira AL (2013) Interleukin-6 gene polymorphism (−174 G/C) is associated with toxoplasmic retinochoroiditis. Acta Ophthalmol 91(4):e311–e314
Cordeiro CA, Moreira PR, Costa GC, Dutra WO, Campos WR, Oréfice F, Teixeira AL (2008) Interleukin-1 gene polymorphisms and toxoplasmic retinochoroiditis. Mol Vis 14:1845–1849
Mesquita RT, Ziegler AP, Hiramoto RM, Vidal JE, Pereira-Chioccola VL (2010) Real-time quantitative PCR in cerebral toxoplasmosis diagnosis of Brazilian human immunodeficiency virus-infected patients. J Med Microbiol 59(Pt 6):641–647
Wujcicka W, Gaj Z, Wilczyński J, Nowakowska D (2015) Possible role of TLR4 and TLR9 SNPs in protection against congenital toxoplasmosis. Eur J Clin Microbiol Infect Dis. doi:10.1007/s10096-015-2461-3
de Sá AR, Moreira PR, Xavier GM, Sampaio I, Kalapothakis E, Dutra WO, Gomez RS (2007) Association of CD14, IL1B, IL6, IL10 and TNFA functional gene polymorphisms with symptomatic dental abscesses. Int Endod J 40(7):563–572
Gorgi Y, Sfar I, Goucha R, Aouadi H, Amri M, Makhlouf M, Ben Romdhane T, Cherif M, Jendoubi-Ayed S, Ben Abdallah T, Ayed K (2010) IL1/IL1 Ra, CTLA-4 and Apo1/Fas genes polymorphisms and susceptibility to IgA nephropathy in Tunisian patients. Tunis Med 88(11):789–793
Klein W, Tromm A, Griga T, Fricke H, Folwaczny C, Hocke M, Eitner K, Marx M, Epplen JT (2001) The polymorphism at position −174 of the IL-6 gene is not associated with inflammatory bowel disease. Eur J Gastroenterol Hepatol 13(1):45–47
Li N, He Z, Xu J, Liu F, Deng S, Zhang H (2010) Association of PDE4D and IL-1 gene polymorphism with ischemic stroke in a Han Chinese population. Brain Res Bull 81(1):38–42
Doyle WJ, Casselbrant ML, Li-Korotky HS, Doyle AP, Lo CY, Turner R, Cohen S (2010) The interleukin 6 –174 C/C genotype predicts greater rhinovirus illness. J Infect Dis 201(2):199–206
Patel JA, Nair S, Ochoa EE, Huda R, Roberts NJ, Chonmaitree T (2010) Interleukin-6−174 and tumor necrosis factor alpha−308 polymorphisms enhance cytokine production by human macrophages exposed to respiratory viruses. J Interferon Cytokine Res 30(12):917–921
Castellucci L, Menezes E, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, Ribeiro S, Reale J, Noronha EF, Wilson ME, Duggal P, Beaty TH, Jeronimo S, Jamieson SE, Bales A, Blackwell JM, de Jesus AR, Carvalho EM (2006) IL6 –174 G/C promoter polymorphism influences susceptibility to mucosal but not localized cutaneous leishmaniasis in Brazil. J Infect Dis 194(4):519–527
Schaaf B, Rupp J, Müller-Steinhardt M, Kruse J, Boehmke F, Maass M, Zabel P, Dalhoff K (2005) The interleukin-6 -174 promoter polymorphism is associated with extrapulmonary bacterial dissemination in Streptococcus pneumoniae infection. Cytokine 31(4):324–328
Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P (1998) The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 102(7):1369–1376
Terry CF, Loukaci V, Green FR (2000) Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem 275(24):18138–18144
Nagineni CN, Detrick B, Hooks JJ (2000) Toxoplasma gondii infection induces gene expression and secretion of interleukin 1 (IL-1), IL-6, granulocyte-macrophage colony-stimulating factor, and intercellular adhesion molecule 1 by human retinal pigment epithelial cells. Infect Immun 68(1):407–410
Acknowledgments
Sponsor: the Polish Ministry of Science and Higher Education, Polish Mother’s Memorial Hospital—Research Institute, grant supporting statutory research no. 2014/II/3.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wujcicka, W., Gaj, Z., Wilczyński, J. et al. Contribution of IL6 −174 G>C and IL1B +3954 C>T polymorphisms to congenital infection with Toxoplasma gondii . Eur J Clin Microbiol Infect Dis 34, 2287–2294 (2015). https://doi.org/10.1007/s10096-015-2481-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-015-2481-z