Abstract
Influenza virus is a major cause of disease worldwide. The accurate detection and further subtyping of influenza A viruses are important for epidemiologic surveillance, and subsequent comprehensive characterization of circulating influenza viruses is essential for the selection of an optimal vaccine composition. ResPlex III is a new multiplex reverse transcriptase polymerase chain reaction (RT-PCR)-based method for detecting, typing, and subtyping influenza virus in clinical specimens. The ResPlex III assay was compared with other methods with respect to sensitivity and accuracy, using 450 clinical specimens obtained from subjects throughout Germany during the 2006–2007 influenza season. Samples were analyzed for the presence of influenza virus in Madin-Darby canine kidney (MDCK) cells by rapid cell culture using peroxidase staining and conventional cell culture confirmed by hemagglutination inhibition assay, a rapid diagnostic assay (Directigen Flu A+B test; BD Diagnostic Systems, Heidelberg, Germany), in-house real-time RT-PCR (RRT-PCR), and ResPlex III (Qiagen, Hilden, Germany). ResPlex III had the highest sensitivity for detecting influenza virus in clinical specimens, followed by in-house RRT-PCR (96% compared with ResPlex III). Conventional cell culture in MDCK cells, rapid culture, and quick test assays were substantially less sensitive (55%, 72%, and 39%, respectively). Virus subtyping results were identical using ResPlex III and the standard virological subtyping method, hemagglutination inhibition. ResPlex III is a quick, accurate, and sensitive assay for detecting and typing influenza A and B viruses and subtyping influenza A viruses in clinical specimens, and might be considered for a supplemental role in worldwide seasonal and pandemic influenza surveillance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Seasonal influenza is responsible for a high disease burden worldwide. Up to 5–15% of the global population is affected by influenza annually [21]. Influenza epidemics result in an average of 294,000 hospitalizations and 36,000 deaths each year in the US alone [15, 16]. The best way to prevent influenza is yearly vaccination with seasonal vaccines that are designed to include antigens from three strains recommended by the World Health Organization (WHO). Virological surveillance to identify predominant circulating influenza virus strains is essential to determine the optimal strains as seasonal influenza vaccine components, and to provide information about the possible emergence of new virus strains with pandemic potential [6]. If vaccine strains do not closely match the strains causing disease in a given year, vaccine efficacy may be reduced, with a potentially significant impact on public health [2].
Reverse transcriptase polymerase chain reaction (RT-PCR)-based methods have replaced viral culture as the reference standard for the detection of influenza viruses [10, 12]. Results are obtained in 2–4 h [3] and, compared to viral culture methods, the specificity is improved and the sensitivity is 2–13% higher [22]. Due to its speed, sensitivity, and specificity, real-time RT-PCR (RRT-PCR) has been recognized as an attractive method of detecting influenza viruses. However singleplex, real-time, one-step RT-PCR cannot distinguish type A and B viruses or allow further subtyping of type A viruses. Thus, different assays have to be used or combined into multiplex PCR assays. New technologies that provide a quicker method of typing and subtyping could enhance global surveillance programs, and more rapid identification of potential pandemic strains would allow more time to prepare for severe outbreaks of disease. These new technologies would need to be at least as sensitive and accurate as existing methods to ensure the continued precision of global surveillance programs. Therefore, the new ResPlex III assay, which detects HA/NA subtypes (i.e., H1 seasonal, H2, H3, H5, H7, H9, N1, and N2) and two influenza A or B generic targets of the non-structural protein (NS) gene, was compared with current diagnostic assays and conventional subtyping by using specific immune sera. Molecular assays are valuable tools for the rapid detection, typing, and subtyping of influenza viruses. However, global influenza surveillance also requires the isolation of virus in culture to allow for a comprehensive analysis of the antigenic profile of circulating influenza viruses, which is important for an optimal vaccine composition.
Study design
Objectives
The aim of this study was to compare the ResPlex III assay with other methods with respect to sensitivity and accuracy, using 450 clinical specimens obtained from subjects throughout Germany during the 2006–2007 influenza season.
Ethical standards
The study was carried out in compliance with the Helsinki Declaration. None of the samples were collected for study purposes. All samples were tested anonymously. Ethical approval was not required.
Study collection
Throat or nasal swabs were obtained from 450 outpatients with acute respiratory symptoms during the 2006–2007 influenza season by office-based physicians, mainly pediatricians and general practitioners in the southern region of Germany. Samples were collected during calendar weeks 4 through 14 of 2007 and stored in virus transport media for routine diagnostics.
Study regime
Rapid test, conventional, and rapid cell culture with adherent Madin-Darby canine kidney (MDCK) cells, nucleic acid extraction, and RT-PCR were performed at the Laboratory Prof. Enders & Partner, Stuttgart, Germany. Aliquots of the extracted nucleic acid were sent to Novartis Vaccines & Diagnostics GmbH for ResPlex III testing. Conventional cell culture isolates were submitted to the National Influenza Reference Center for Influenza for typing/subtyping by hemagglutination inhibition (HI). In addition, RRT-PCR-negative but ResPlex III-positive clinical specimens were used for the isolation of influenza viruses in MDCK33016PF suspension culture developed by Novartis Vaccines & Diagnostics GmbH [9].
BD Directigen Flu A+B (rapid test)
The BD Directigen Flu A+B is a rapid membrane enzyme immunoassay, carried out with fresh clinical specimens according to the manufacturer’s instructions (Becton Dickinson, Sparks, MD).
Conventional cell culture
MDCK cells were grown in 3-mL flat Nunclon tubes (Nunc, Langenselbold, Germany) in Eagle’s Minimal Essential Medium (EMEM) containing 10% fetal bovine serum (FBS). Prior to infection, the medium was replaced with 2 mL of maintenance medium without FBS containing tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin (2 μg/mL). Cell culture tubes were inoculated with 0.2 mL of samples and incubated at 37°C. After 24 h, the medium was replaced by 2 mL of maintenance medium without FBS containing TPCK trypsin (2 μg/mL). The tubes were checked daily for 7 to 10 days for cytopathic effects (CPEs). After 7 to 10 days, the tubes were screened by hemagglutination assay for the agglutination of guinea pig erythrocytes (Charles River Laboratories, Sulzfeld, Germany) in the tissue culture supernatant to confirm influenza virus replication.
Hemagglutination assays (adherent MDCK)
Hemagglutination assays were performed in microtiter U-bottom plates with 25 μL of culture supernatant, 25 μL of dextrose-gelatin-veronal (DGV) buffer, and 25 μL of washed 0.5% guinea pig erythrocytes (Charles River Laboratories, Sulzfeld, Germany), suspended in DGV buffer. The plates were incubated at room temperature for approximately 1 h.
HI assays
The HI procedure for subtyping influenza virus isolates has been described previously [4]. Briefly, specific antisera raised in ferrets were treated with receptor-destroying enzyme. HI assays were performed using 4 hemagglutination units of virus and 0.75% (vol/vol) guinea pig erythrocytes.
Rapid culture
The rapid culture technique was performed as described previously [14]. MDCK cells were seeded in 96-well microtiter plates in antibiotic-free EMEM containing 10% FBS and incubated at 37°C for 1–3 days before testing. For the detection of influenza A and B viruses in clinical specimens, two wells were each inoculated with 100 μL of sample after removing the medium. Plates were centrifuged at 1,200 × g for 30 min. The supernatant was then removed and 200 μL of FBS-free medium containing TPCK trypsin (2 μg/mL) were added. Plates were incubated in a moist chamber at 37°C for 16–18 h under 5% CO2. Subsequently, the cells were fixed for 10 min with ice-cold acetone/methanol (40:60), then blocked with 1% bovine serum albumin (BSA) in phosphate buffered saline (PBS) for 30 min, and incubated at 37°C for 30 min with monoclonal antibodies produced in mice against influenza A or B (Chemicon, Temecula, CA) diluted 1:100 in blocking buffer (1% BSA in PBS). After washing three times with PBS, the plates were incubated with a secondary horseradish peroxidase-labeled antibody (DAKO, Glostrup, Denmark) at a dilution of 1:500 in blocking buffer. The washing step was repeated. Influenza-positive cells were stained using the substrate 3-amino-9-ethylcarbazole (AEC) (Sigma, St. Louis, MO) and counted microscopically.
MDCK 33016PF suspension culture
MDCK 33016PF, where PF indicates “protein free” and 33016 is the clone number, is a proprietary cell line that was generated by Novartis [9]. The MDCK 33016PF cells from Novartis’ working seed stocks were cultivated in 500-mL spinner flasks (Wheaton) in CDM medium (MDCK 33016 CDM, Lonza) and passaged in 3–4-day intervals. During those 3–4 days, the cells grew from an initial seeding density of 1 × 105 cells/mL to densities between 1.0 and 1.5 × 106 cells/mL. For infections, 4.5 mL of cells were seeded in 50-mL filter tubes (TPP, Trasadingen, Switzerland) at a cell density of 0.8 to 1.2 × 106 cells/mL in infection medium, consisting of 70% MDCK 33016 PFM medium (Gibco Invitrogen) and 30% MDCK 33016 CDM supplemented with 0.5% of a penicillin/streptomycin solution (Sigma) and 900 IU/mL trypsin. To obtain a total culture volume of 5 mL, the added inoculum clinical (stored at 2–8°C) or virus sample was diluted in 0.5 mL of infection medium and was prediluted by several log10 steps, starting with a dilution of at least 1:5. Inoculated cultures were then incubated at 33°C for 3 days in a 5% CO2 atmosphere in an ISF-1-W shaker incubator (Kühner, Birsfelden, Switzerland). For virus harvests, the cells were separated by centrifugation (800–1,000 × g for 10 min) and the supernatant was recovered. Unless used freshly (e.g., for hemagglutination tests and subsequent passaging), aliquots of the supernatant were frozen at ≤−60°C.
Hemagglutination assays (MDCK 33016PF)
Hemagglutination (HA) testing was done with harvested material to define the starting material for the next passage. HA testing was performed in U-bottom microwell plates (Greiner Bio-One) using 100 μL of a serial log2 dilution in PBS (pH 7.0) of the test samples and 100 μL of chicken or guinea pig red blood cells (0.5% in PBS, pH 7.0). The results were read after 60 min (chicken erythrocytes) or 120 min (guinea pig erythrocytes) incubation at ambient temperature within a temperature range of 19–25°C. Two different kinds of red blood cells were used, since H3N2 influenza strains did not react with chicken red blood cells. Material from the highest log10 inoculum dilution, which showed a clearly positive HA reaction after the previous passage, was used for the following passage.
Automated nucleic acid isolation
Viral RNA was prepared from up to 32 samples in parallel with the automated MagNA Pure instrument by using the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche Diagnostics GmbH, Mannheim, Germany). Briefly, the input sample volume was 200 μL and nucleic acids were eluted in a volume of 50 μL. Purified RNA was stored at −20°C.
RT-PCR
The duplex RRT-PCR was done using the LightCycler system. Amplification was performed in a 20-μL reaction volume consisting of 10 μL of kit-supplied QuantiTect Probe Master Mix (Qiagen, Hilden, Germany), 0.5 μM of each primer, 0.18 μM of probe, 0.2 μL of kit-supplied QuantiTect RT mix, and 6.8 μL of purified RNA. Primers and the two single probes were described previously [19]. Duplex real-time PCR was carried out with an initial RT step at 50°C for 20 min, followed by PCR activation at 95°C for 15 min and 50 cycles of amplification (95°C for 5 s, 55°C for 20 s, 72°C for 30 s). Fluorescence development was measured once each cycle after the elongation step.
ResPlex III
An aliquot of the viral RNA which was used for in-house RT-PCR was shipped on dry ice to Novartis, Marburg, Germany. RT-PCR was performed using the OneStep RT-PCR Kit (Qiagen, Hilden, Germany). The reaction mixture included 5 μL of nucleic acids, 10 μL of 5x Qiagen OneStep RT-PCR buffer, 2 μL of dNTP mix, 6 μL of ResPlex III primer mix, 2 μL of Qiagen OneStep RT-PCR enzyme mix, and 25 μL of RNase-free water. Amplification was performed on a Mastercycler ep gradient S thermocycler (Eppendorf, Hamburg, Germany), with the ramp rate set to 15%. RT-PCR was carried out with an initial RT step at 50°C for 30 min, followed by 15 min of PCR activation at 95°C, 15 cycles of enrichment cycling (94°C for 30 s, 52°C for 1 min, 72°C for 1 min), six cycles of two-step cycling (94°C for 15 s, 70°C for 1.5 min), and 30 cycles of three-step cycling (94°C for 15 s, 52°C for 15 s, 72°C for 15 s). A final extension of 3 min at 72°C concluded the amplification.
Following thermocycling, 5 μL of RT-PCR product, 10 μL of ResPlex III Bead Mix, and 35 μL of detection buffer were incubated at 52°C for 10 min. For detection, 10 μL of streptavidin–phycoerythrin conjugate mixed with detection buffer were added. Mixtures were maintained at 52°C for 5 min, and then 120 μL of stopping buffer was added. Samples were analyzed on a Luminex 200 system (Luminex, Austin, TX) running QIAplex MDD Software 1.1.15. Raw mean fluorescence intensity data were exported to Excel for storage and analysis. One negative control (RNase-free water) and one independent positive control were processed in parallel for each analysis.
Determining detection limits
Laboratory-grown influenza virus stocks of A/Bayern/7/95 (H1N1) and B/Baden Württemberg/3/06 with titers of 8.1 and 7.9 TCID50/mL, respectively, were diluted 108-fold in ten-fold increments in virus transport medium. These serial dilutions were used to establish the relationship between influenza viral load determined by ResPlex III or by RRT-PCR (i.e., between TCID50/mL and viral cDNA copies), since there is no international standard for influenza A and B viral RNA. To determine TCID50, the influenza A and B viruses were serially diluted ten-fold. Eight wells of a 96-well plate containing a confluent monolayer of MDCK cells (MDCK 33016 grown adherently) were inoculated with 0.1 mL of each dilution, and the cells were incubated at 37.0°C in a humidified atmosphere under 5.0% CO2. After 5 days, the cell cultures were examined microscopically for CPE. The virus titer was calculated using the Spearman–Karber method [8, 13].
Results
Demographics
Subjects were evenly split between genders (49% female, n = 222; 51% male, n = 228). Since most of the referring doctors were pediatricians, the majority of clinical isolates came from children, although the subject ages ranged overall from 0 to 76.5 years (Fig. 1).
Sensitivity
For RRT-PCR, the detection limits were 2.1 and 2.9 log10 TCID50/mL for A/H1N1 and B, respectively. ResPlex III showed higher sensitivity for the NS gene target than for the HA gene (H1) or NA gene (N1). The detection limits for the NS gene target were 1.1 and 1.9 log10 TCID50/mL for A/H1N1 and B, respectively. Thus, ResPlex III was at least one log more sensitive than in-house RRT-PCR.
Detection of type A and B viruses
Five assays were performed on each sample: ResPlex III, RRT-PCR, rapid cell culture, conventional cell culture, and a rapid diagnostic test (BD Directigen Flu A+B). ResPlex III identified the greatest number of positive isolates (n = 261, or 58%). Thus, the sensitivity of ResPlex III was set at 100% and all other assays were calculated as a subset of that total (Table 1). The sensitivity of RRT-PCR was 96%. Cell culture techniques and the rapid test were far less sensitive than molecular techniques, identifying 39% (BD Directigen Flu A+B test) to 72% (rapid cell culture) of influenza-positive specimens. One sample tested influenza A-positive using the BD Directigen Flu A+B test and negative using all other assays, suggesting a false-positive result.
One patient sample contained both A/H3N2 and B viruses. Although the in-house RRT-PCR can distinguish influenza A from influenza B, only the ResPlex III assay was able to identify this dual infection and subtype at the same time. Cell-based methods identified only the B virus, and the BD rapid test produced a negative result.
Only nine samples were positive for influenza B virus in this study (using ResPlex III). In this limited comparison, the sensitivity of the other assays for detecting influenza B virus appeared to be lower than the sensitivity for detecting influenza A, and the molecular methods showed greater sensitivity for detecting influenza B than the other methods. The in-house RRT-PCR method detected 44% of samples that were identified as influenza B-positive with ResPlex III. Analysis of the cycle thresholds (Ct, the number of PCR cycles required to amplify nucleic acid above background levels) confirmed that ResPlex III was more sensitive than RRT-PCR, particularly for the detection of influenza B virus. Conventional cell culture and rapid culture detected only three and one of these samples, respectively. No sample was influenza B-positive with the BD Directigen Flu A+B test.
Sensitivity of cell culture and rapid diagnostic methods in different age groups
Particularly in adults, the two cell culture methods and the rapid test showed different sensitivities (Table 2). In one out of two cases, influenza could be diagnosed in children using the BD Directigen Flu A+B test. However, in this study, the sensitivity of the rapid test dropped in specimens of adults and the elderly to 14.3%. The same result was found using rapid cell culture. Interestingly, the sensitivity of the conventional cell culture method (virus isolation on MDCK cells) remains unchanged (about 50%) in all of the age groups.
Typing efficiency of ResPlex III depends on viral load
The analysis of Ct values confirmed that ResPlex III was more sensitive than the RRT-PCR, particularly for the B virus (Table 3). The Ct value is the number of PCR cycles required to amplify nucleic acid above background levels based on the intensity of fluorescently labeled products. Therefore, a higher Ct value indicates a lower starting quantity of nucleic acid. In this study, there were several samples with very high Ct values that had definitively positive ResPlex III results. Also, ten samples (out of 261, or 3.85%) tested positive using the ResPlex III assay but tested negative with RT-PCR. These ten samples which were tested positive in ResPlex III but tested negative in RRT-PCR were subjected to the MDCK 33016PF suspension culture for virus isolation. Five of the ten samples were tested positive after the second passage in the hemagglutination assay, confirming that, for these samples, the ResPlex III results were true-positive: from one of three H3N2-positive and four of five B-positive probes, influenza viruses were cultivated. In addition, one B-positive material was also cultivated in adherent MDCK cells and subtyped by the Robert Koch Institute (RKI).
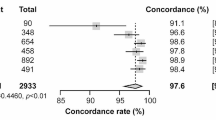
Specificity of typing/subtyping by ResPlex III
HI assays were performed on 145 MDCK isolates to determine the accuracy of ResPlex III subtyping (Table 4). In all, HI serotyping was accomplished on 135 (93%) samples; 134 (99%) of these were identical to the subtypes found with ResPlex III. Nine virus isolates could not be further passaged or grew with too low HA titers, not allowing HI testing. The one sample that was not identical contained both A/H3N2 and B according to ResPlex III, but only B was isolated in conventional cell culture and, subsequently, also identified by HI. Thus, ResPlex III was able to identify viral types/subtypes as accurately as HI typing and subtyping of viruses isolated from PCR-positive specimens. To further analyze samples that tested weakly positive by ResPlex III and negative by RRT-PCR, Novartis MDCK 33016PF suspension cells were used for influenza virus recovery. Six out of these 11 specimens were positive by hemagglutination assays after the first or second passage. In conclusion, these weak-positive results obtained by ResPlex III were correct and could be confirmed by some positive virus isolations in the Novartis MDCK 33016PF suspension cell line.
Comparison to national and global surveillance data
Data from the WHO were collected from an online database (FluNet) to determine the epidemiological data from Europe [23]. National influenza surveillance in Germany is performed by the Working Group on Influenza (“Arbeitsgemeinschaft Influenza”, AGI) at the RKI [25]. Here, AGI data from Germany [1] and WHO data from Europe (week 4 to week 14 of 2007) were compared with the results from this study.
The proportion of samples from patients with respiratory symptoms that were positive for influenza virus varied from 23 to 58% in this study, depending on the assay used. Rates of influenza virus detection were 58 and 56% for ResPlex III and RRT-PCR, respectively, and were comparable to data reported for national influenza surveillance by the AGI and WHO that reported a detection rate of 53 and 25% for the same time period and geographic region (Fig. 2a).
a Comparison of influenza-positive samples determined by real-time reverse transcriptase polymerase chain reaction (RRT-PCR) in this study to Arbeitsgemeinschaft Influenza (AGI) and World Health Organization (WHO) data. The percentage of influenza-positive samples obtained in this study was similar to those found during the 2006–2007 season by local and regional influenza surveillance systems (AGI in Germany and the WHO in Europe). However, the greater sensitivity of molecular techniques may contribute to the higher values seen for ResPlex III and RRT-PCR. b Comparison of influenza virus subtyping results to the AGI and WHO data. When influenza-positive samples are sorted according to subtype, the relative proportion of each subtype is similar for ResPlex III and the data from Europe (obtained by the WHO) and from Germany (obtained by the AGI) (black = influenza B, gray = influenza A, blue = A/H1N1, green = A/H3N2, red = A/H3N2 + B)
The influenza virus subtyping results obtained with ResPlex III in the present study were also similar to the data reported by the AGI and the WHO. During the 2006–2007 influenza season, the National Influenza Reference Center characterized 1,130 influenza virus isolates in Germany and, according to the WHO data, 9,024 positive samples were identified in Europe, of which 85% and 47% were A/H3N2, 14% and 5% were A/H1N1, and 2% and 4% were influenza B, respectively. The WHO also reported on 3,996 influenza A viruses, which were not subtyped, accounting for 44% of all influenza-positive samples. In this study, the relative proportions of these strains were similar to data of the national influenza surveillance and the European WHO data (Fig. 2b). ResPlex III identified 78% of samples as containing A/H3N2, 12% as A/H1N1, 3% as influenza B, 7% as untyped influenza A, and <1% as co-infected with A/H3N2 and influenza B. Of the samples that were untyped influenza A, all 17 were NS gene-positive, but ten were negative for either the HA or NA genes and seven were negative for both the HA and NA genes.
Discussion
Since only a vaccine whose virus strains match the circulating influenza viruses will protect vaccinees efficiently, frequent updating of the influenza vaccine composition is necessary. Therefore, the surveillance of influenza virus strains that are circulating regionally and globally is essential and requires highly sensitive and accurate diagnostic tests. Using isolates collected during the 2006–2007 influenza season, we have shown that ResPlex III can detect and subtype strains of influenza A viruses with adequate sensitivity and accuracy.
Influenza B viruses co-circulate along with influenza A strains and cause epidemics in some years [6]. The sensitivity of several diagnostic tests for detecting influenza B virus has been shown to be less than for the influenza A virus [7]. In this study, only nine influenza B virus infections could be detected. Since there were relatively few samples containing influenza B virus in this study, no conclusion could to be drawn about a higher sensitivity of ResPlex III compared to the other methods. However, the sensitivity of the in-house real-time PCR was improved in further seasons after the QuantiTect RT mix from Qiagen was replaced by the QuantiTect multiplex RT-PCR master mix.
Among the analyzed samples, one sample contained both A/H3N2 and B according to ResPlex III, but only B was isolated in conventional cell culture and, subsequently, also identified by HI. However, this difference is most likely due to a limitation of cell-based assays: even when multiple viral subtypes infect a cell, usually, only one subtype persists and replicates [5, 17, 18].
In the present study, the sensitivities of conventional cell culture, rapid culture, and quick test assays proved already to be substantially lower than that of RRT-PCR and ResPlex III. A study with specimens from adults and the elderly would probably have yielded even lower sensitivities for these other detection methods. As shown in Table 3, the positive detection rate of rapid cell culture and rapid test (BD Directigen Flu A+B) was particularly high in kindergarten and school-aged children. In this study, clinical samples of children were overrepresented (Fig. 1), because children aged <5 years have more influenza-related medical care visits compared with older children, and, particularly, those aged <2 years are at the greatest risk for influenza-related hospitalizations [6, 14].
ResPlex III is an assay that is available as a customized product for detecting and subtyping influenza virus in clinical samples. Due to its multiplex format, ResPlex III can detect more than one viral template in the same reaction mixture, allowing viral co-infections to be identified in the same specimen. In addition, ResPlex III as well other specific molecular assays provide subtyping results within a few hours compared with about one week required for cell culture isolation and subsequent differentiation techniques. However, the high sensitivity and subtyping accuracy of ResPlex III that was demonstrated in this study should be confirmed by analyzing more strains including H1N1 (2009) viruses collected from different seasons.
ResPlex III is able to detect subtypes with pandemic potential (such as H2, H5, H7, and H9) that are not normally screened in routine diagnostic procedures [24]. To maintain the usefulness of the ResPlex III assay in influenza surveillance, regular updates are important to ensure that currently circulating influenza A subtypes as well as other potentially harmful subtypes could be detected. For example, the current ResPlex III panel could be enhanced by the addition of the H4 and H6 subtypes, which may have pandemic potential [20]. In general, this technology allows the fast addition of new subtypes to the panel. This was the case with the A/H1N1 (2009) strain, which was implemented in only a few weeks in the ResPlex II Plus Panel RUO [11], and, thus, the ResPlex III panel should also be updated rapidly by including the specific detection of the H1N1 (2009) virus.
In conclusion, the use of ResPlex III might potentially translate into a quicker response to a potential pandemic threat and provide timelier subtyping of circulating influenza A strains during seasonal outbreaks. Thus, ResPlex III has potential benefits for monitoring circulating influenza viruses as a supplemental assay for influenza surveillance.
References
Arbeitsgemeinschaft Influenza (AGI) Saisonabschlussbericht der Arbeitsgemeinschaft Influenza 2006/2007. Available online at: http://www.influenza.rki.de/Saisonberichte/2006.pdf. Accessed 2 February 2011
Carrat F, Flahault A (2007) Influenza vaccine: the challenge of antigenic drift. Vaccine 25:6852–6862
Centers for Disease Control and Prevention (CDC) (2010) Influenza symptoms and laboratory diagnostic procedures. Available online at: http://www.cdc.gov/flu/professionals/diagnosis/labprocedures.htm. Accessed 21 October 2008
Chakraverty P (1971) Antigenic relationship between influenza B viruses. Bull World Health Organ 45:755–766
Falchi A, Arena C, Andreoletti L, Jacques J, Leveque N, Blanchon T, Lina B, Turbelin C, Dorléans Y, Flahault A, Amoros JP, Spadoni G, Agostini F, Varesi L (2008) Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Corsica Island, France. J Clin Virol 41:148–151
Fiore AE, Shay DK, Broder K, Iskander JK, Uyeki TM, Mootrey G, Bresee JS, Cox NS; Centers for Disease Control and Prevention (CDC); Advisory Committee on Immunization Practices (ACIP) (2008) Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2008. MMWR Recomm Rep 57(RR-7):1–60
Hurt AC, Alexander R, Hibbert J, Deed N, Barr IG (2007) Performance of six influenza rapid tests in detecting human influenza in clinical specimens. J Clin Virol 39:132–135
Karber G (1931) Beitrag zur kollectiven behandlung pharmakologischer reihenversuche. Arch Exp Path Pharmak 162:480–483
Onions D, Egan W, Jarrett R, Novicki D, Gregersen JP (2010) Validation of the safety of MDCK cells as a substrate for the production of a cell-derived influenza vaccine. Biologicals 38:544–551
Petric M, Comanor L, Petti CA (2006) Role of the laboratory in diagnosis of influenza during seasonal epidemics and potential pandemics. J Infect Dis 194(Suppl 2):S98–S110
Qiagen ResPlex II Plus Panel RUO. For multiplex detection of respiratory viral targets. Available online at: http://www1.qiagen.com/Products/ResPlexIIPlusPanelRUO.aspx. Accessed 2 December 2009
Schweiger B, Zadow I, Heckler R, Timm H, Pauli G (2000) Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J Clin Microbiol 38:1552–1558
Spearman C (1908) The method of “right and wrong cases” (constant stimuli) without Gauss’s formulae. Br J Psychol 2:227–242
Terletskaia-Ladwig E, Eggers M, Meier S, Leinmüller M, Schneider F, Schmid M, Enders M (2009) Laboratory-based assessment of influenza in German ambulant patients from 1998 to 2008. Infection 37:401–406
Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K (2004) Influenza-associated hospitalizations in the United States. JAMA 292:1333–1340
Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K (2003) Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186
Toda S, Okamoto R, Nishida T, Nakao T, Yoshikawa M, Suzuki E, Miyamura S (2006) Isolation of influenza A/H3 and B viruses from an influenza patient: confirmation of co-infection by two influenza viruses. Jpn J Infect Dis 59:142–143
Waner JL (1994) Mixed viral infections: detection and management. Clin Microbiol Rev 7:143–151
Ward CL, Dempsey MH, Ring CJ, Kempson RE, Zhang L, Gor D, Snowden BW, Tisdale M (2004) Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol 29:179–188
Webby RJ, Webster RG (2003) Are we ready for pandemic influenza? Science 302:1519–1522
World Health Organization (WHO) (2009) Influenza (seasonal). Fact sheet no. 211. Available online at: http://www.who.int/mediacentre/factsheets/fs211/en. Accessed 13 June 2008
World Health Organization (WHO) (2005) WHO recommendations on the use of rapid testing for influenza diagnosis. Available online at: http://www.who.int/csr/disease/avian_influenza/guidelines/RapidTestInfluenza_web.pdf. Accessed 21 October 2008
World Health Organization (WHO) (2003–2007) Global health atlas. Available online at: http://www.who.int/globalatlas/dataQuery/default.asp. Accessed 21 October 2008
Zou S, Han J, Wen L, Liu Y, Cronin K, Lum SH, Gao L, Dong J, Zhang Y, Guo Y, Shu Y (2007) Human influenza A virus (H5N1) detection by a novel multiplex PCR typing method. J Clin Microbiol 45:1889–1892
Zucs P, Buchholz U, Haas W, Uphoff H (2005) Influenza associated excess mortality in Germany, 1985–2001. Emerg Themes Epidemiol 2:6
Acknowledgments and conflicts of interest
Bernhard Roth and Jens-Peter Gregersen (Novartis Vaccines and Diagnostics) were supported by HHS grant 0100200600012 C from the U.S. Department of Health & Human Services. Maren Eggers, Martin Enders, and Michael Schmid (Laboratory Prof. Enders & Partners) received financial support from Novartis Vaccines and Diagnostics. Brunhilde Schweiger has no conflicting interests to declare. Premarket ResPlex III kits were provided by Qiagen (Hilden, Germany). The excellent technical assistance of Michael Leinmüller, Franz Schneider, Carolin Benzinger, Silvia Meier, Knut Schwarz, and Marion Elsen is gratefully acknowledged. Funding support for medical editorial assistance was provided by Novartis Vaccines and Diagnostics. We thank Pamela Tuttle of Health Interactions (Atlanta, GA) for the medical editorial assistance with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Maren Eggers and Bernhard Roth contributed equally to the work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Eggers, M., Roth, B., Schweiger, B. et al. Comparison of the novel ResPlex III assay and existing techniques for the detection and subtyping of influenza virus during the influenza season 2006–2007. Eur J Clin Microbiol Infect Dis 31, 1257–1265 (2012). https://doi.org/10.1007/s10096-011-1437-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-011-1437-1