Abstract
The development of fast and easy on-site molecular detection and quantification methods for hazardous microbes on solid surfaces is desirable for several applications where specialised laboratory facilities are absent. The quantification of bacterial contamination necessitates the assessment of the efficiency of the used methodology as a whole, including the preceding steps of sampling and sample processing. We used quantitative real-time polymerase chain reaction (qrtPCR) for Escherichia coli and Staphylococcus aureus to measure the recovery of DNA from defined numbers of bacterial cells that were subjected to three different DNA extraction methods: the QIAamp® DNA Mini Kit, Reischl et al.’s method and FTA® Elute. FTA® Elute significantly showed the highest median DNA extraction efficiency of 76.9% for E. coli and 108.9% for S. aureus. The Reischl et al. method and QIAamp® DNA Mini Kit inhibited the E. coli qrtPCR assay with a 10-fold decrease of detectable DNA. None of the methods inhibited the S. aureus qrtPCR assay. The FTA® Elute applicability was demonstrated with swab samples taken from the International Space Station (ISS) interior. Overall, the FTA® Elute method was found to be the most suitable to selected criteria in terms of rapidity, easiness of use, DNA extraction efficiency, toxicity, and transport and storage conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The detection and quantification of microbes present on solid surfaces plays an important role in clinical settings, the food industry, drinking water distribution systems, air-conditioning systems in modern buildings or advanced life-support systems, such as manned spacecraft. Detection and quantification by conventional culturing techniques, however, is labourious, lengthy, requires specialist expertise, has poor diagnostic sensitivity and, in addition, many bacteria such as Faecalibacterium prausnitzii are difficult to culture or uncultivable, which hampers their detection [1]. Moreover, the culturing of hazardous microbes such as pathogens and so-called “technophiles” may often be undesirable [2]. Especially in remote locations such as field hospitals, disaster areas or manned spacecraft, where specialised laboratory facilities and technical personnel are not available, this results in the need for fast and easy methodologies that can be used on-site by non-specialists. Accordingly, molecular methods have increasingly been used for the detection and quantification of hazardous microbes, overcoming many of the disadvantages encountered by culturing [3].
Quantitative real-time polymerase chain reaction (qrtPCR) is a cultivation-independent method for the rapid molecular detection and quantification of nucleic acid sequences [3]. The method has a high specificity with a detection sensitivity of a few molecules per reaction and the potential to be automated and miniaturised for use on-site by non-specialists. Moreover, for a reliable quantification of bacterial contamination, it is important to assess the efficiency of the used methods as a whole, including both the molecular detection method as well as the preceding steps of sampling and sample processing [3–5]. In addition, for increased sensitivity, methods with the highest possible efficiency should be utilised and the dilution factor in all steps of sampling and sample processing kept to a minimum.
Sample processing for use with qrtPCR involves cell lysis and the subsequent recovery of DNA free of amplification inhibitors. Factors that play a role in the efficiency of bacterial cell lysis may be physiological characteristics of the species such as the constitution of the cell wall, the physiological state which the cell is in or cell concentration [6–8]. As a consequence, most DNA extraction methods are optimal for just one or a group of bacterial species [6]. Ideally, for applications where it is essential to measure the levels of a variety of hazardous species, such as in a natural environment, a universal DNA extraction method would be preferred that is optimal for all bacterial species.
The aim of this study was to select and validate a suitable DNA extraction method for the recovery of DNA from the potential pathogens Escherichia coli and Staphylococcus aureus to be used in qrtPCR. These species were used as model organisms, as the Gram-negative bacterium E. coli lyses differently compared to the Gram-positive bacterium S. aureus due to a different constitution of its cell wall. In particular for use on-site with samples taken from solid surfaces, we sought to select a fast and easy-to-use DNA extraction method that has a high efficiency, is universally effective for bacterial species, makes use of non-toxic chemicals and has little or no transport or storage requirements.
Materials and methods
Cultivation of bacterial cells
S. aureus subsp. aureus ATCC 25923 and E. coli ATCC 11775T cells were each inoculated into 9 ml Brain Heart Infusion (BHI) (Media Products BV, Groningen, The Netherlands) and grown overnight at 37°C. From each culture, cells were washed twice by centrifugation of 1 ml of cell suspension for 10 min at 16,100g, removal of the supernatant and resuspension of the pellet in 1 ml of sterile physiological salt solution (0.85% NaCl, Media Products BV, Groningen, The Netherlands). The suspensions were kept on ice during the remainder of the procedure to minimise cell growth and lysis.
DNA extraction methods description
With the commonly used QIAamp® DNA Mini Kit (QIAGEN GmbH, Hilden, Germany), total DNA can be purified from a variety of biological, clinical and forensic specimens. The method uses chemical lysis by undisclosed lysis buffers containing chaotropic salt, enzymatic digestion by treatment with proteinase K and thermal lysis. The method uses a separate protocol for Gram-positive bacteria, with the additional use of enzymatic digestion by lysozyme or lysostaphin with Triton. DNA is bound to a silica-gel membrane in a spin-column, whilst PCR inhibitors are, supposedly, not retained. The bound DNA is then washed, eluted in buffer and can then be stored at −20°C.
The simple boiling procedure developed by Reischl et al. involves chemical lysis in combination with thermal lysis and has been used for the extraction of S. aureus total DNA [9]. The concentrated lysis buffer (10×) contains 10% Triton X-100, 5% Tween 20, 100 mmol l−1 Tris-HCl (pH 8.0) and 10 mmol l-1 EDTA.
The FTA® Elute method (Whatman plc., Maidstone, UK) was developed for room-temperature collection, transport, storage and isolation of nucleic acids from a wide variety of biological, clinical and forensic sample types. The method uses chemical lysis by a cellulose paper matrix treated with proprietary reagents that lyses cells and denatures proteins upon contact, causing the release and subsequent entrapment of nucleic acids in the fibres of the matrix. The paper rapidly inactivates organisms, decreasing the risk of contamination for the individuals handling the sample. Sample volume requirements are minimal, approximately 12–40 μl per collection area. After drying, the samples can be stored for many years. DNA is recovered from the FTA® Elute matrix through a simple hot water elution procedure. Inhibitory components are supposedly retained on the FTA® Elute matrix or disposed of during a short washing step.
DNA extraction method comparison
The experiments were performed using DNA-free laboratory techniques. The cells in both E. coli and S. aureus suspensions were enumerated by 4′,6-diamidino-2-phenylindole (DAPI) staining and subsequent visual cell counting with an Olympus BH2 epifluorescence microscope by the method as described previously [10], with the exception that the cells were not fixed by paraformaldehyde. The enumerations were performed in duplicate and averaged. Subsequently, the cell suspensions were serially diluted in sterile physiological salt solution and divided into aliquots of 5.3 × 105, 5.3 × 104, 5.3 × 103, 5.3 × 102 and 5.3 × 101 cells.
The volumes of the aliquots were adjusted with sterile physiological salt solution to suit the recommended input volume for each DNA extraction method, after which DNA was extracted in quadruplicate according to the instructions of the authors and manufacturers and subjected to qrtPCR as described below.
To monitor contamination during the procedure, negative (extraction) controls were prepared consisting of physiological salt solution processed through each DNA extraction method. PCR inhibition was determined by adding a template of 1 ng of the qrtPCR standard DNA, prepared as described below, together with either 2 μl of an extraction control or 2 μl Molecular Biology Water to triplicate reactions. PCR inhibition will cause a shift of the threshold cycle (Ct) to a higher cycle number compared to the standard (Online Resources 1 and 2). Positive controls were performed by adding a template of 100 fg of the standard together with 2 μl Molecular Biology Water to reactions in triplicate.
DNA extraction of swab samples taken from the International Space Station interior using the FTA® Elute method
To demonstrate the applicability of the FTA® Elute method for the analysis of surface samples, DNA was extracted with this method from triplicate swab samples taken from the interior of the International Space Station (ISS) as part of the “SAMPLE” experiments, which were previously described by van Tongeren et al. in 2007 [2]. The DNA contents were subsequently quantified with the qrtPCR assays of S. aureus and E. coli as described below.
Quantitative real-time PCR methods
Two TaqMan PCR assays were used to amplify specific regions of the genomes of S. aureus and E. coli. Oligonucleotide primers and double-dye probes were obtained from Eurogentec S.A. (Seraing, Belgium). Primers SauV58 (5′-GCTGTGATGGGGAGAAGACAT-3′) and SauR54b (5′-CGGTACGGGCACCTATTTTC-3′) were used to amplify a 90-bp fragment of the 23S rRNA gene of S. aureus [7]. Primers UIDA-F (5′-TGGTGATTACCGACGAAAAC-3′) and UIDA-R (5′-GCGTGGTTACAGTCTTGC-3′) were used to amplify a 145-bp fragment of the uidA gene of E. coli [11]. Probes Sta59bTQ (5′-AGAGGCTTTTCTCGGCAGTGTGAAATCAACGA-3′) and UIDA-Pr (5′-GCCGGGATCCATCGCAGCGTAATGCTC-3′) were hybridised in real-time with the PCR product of S. aureus and E. coli, respectively [7, 11]. The probes were labelled with carboxyfluorescein (FAM) on the 5′-end and a 4-([4-(dimethylamino)phenyl]azo)benzoic acid (DABCYL) quencher on the 3′-end. All qrtPCR reactions were performed in a total volume of 30 μl, consisting of 3 μl of target DNA and 27 μl of amplification mixture containing PCR Reaction Buffer (Smart™ Kit No ROX, Eurogentec S.A.), primers, probe and Molecular Biology Water (AccuGENE®, Lonza Group Ltd., Basel, Switzerland). The Smart™ Kit PCR reagents contained HotGoldStar DNA polymerase and uracil-N-glycosylase to prevent carry-over contamination. The end concentration of each primer was 150 nmol l−1, of the Sta59bTQ probe 80 nmol l−1 and of the UIDA-Pr probe 300 nmol l−1. Amplification and detection were carried out on a Smart Cycler® System (Cepheid, Sunnyvale, CA, USA) with a profile of 50°C for 2 min to activate uracil-N-glycosylase, 95°C for 10 min to activate HotGoldStar DNA polymerase, followed by 45 cycles of 95°C for 15 s and 60°C for 60 s.
Negative controls for each qrtPCR assay were performed by running reactions without the addition of template DNA.
Generation of qrtPCR standards with bacterial genomic DNA
Cells were cultivated as described above. To obtain a sufficient amount of relatively good quality DNA, the commonly used QIAamp® DNA Mini Kit was used to extract DNA from the cells according to the instructions of the manufacturer. The procedure was followed by ethanol precipitation to purify the DNA solution from contaminants such as PCR-inhibiting substances. The DNA purity and concentration was determined with a NanoDrop® ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). A standard curve of mean qrtPCR Ct values was prepared with triplicate replicates of serial dilutions of bacterial genomic DNA (three series of 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg and 10 fg for S. aureus subsp. aureus ATCC 25923 and two series of 30 ng, 3 ng, 300 pg, 30 pg, 3 pg, 300 fg and 30 fg for E. coli ATCC 11775T, respectively). The amount of bacterial DNA in samples that were subjected to the various DNA extraction methods was estimated by qrtPCR with this curve under identical PCR conditions.
Statistical analysis
The quantification limit was determined for each qrtPCR assay as the lowest concentration on the linear part of the standard curve at which 7 out of 9 reactions or more gave a positive result. The quantification limit was used as the cut-off level or, instead of this, the value of a negative control in case it had a positive result. In case a Ct of zero (negative result) or above the mean Ct (+ 1 s.d.) of the quantification limit was measured, a Ct equal to the mean Ct (+ 1 s.d.) of the quantification limit was assumed for the data analysis.
DNA extractions were performed in quadruplicate. The efficiency of DNA extraction was determined by the measured amount of bacterial DNA in each sample as compared to the estimated amount of bacterial DNA before DNA extraction as a reference (100%). The reference was estimated by microscopic cell counting and assuming a chromosomal weight calculated from the chromosome length and GC content of approximately 3.0 fg for S. aureus and 5.7 fg for E. coli, respectively. Within each DNA extraction method, the efficiencies of extraction amongst different cell amounts were compared by Kruskal–Wallis analysis. Friedman analysis with additional Bonferroni-protected contrasts was performed to compare DNA extraction methods amongst each species. Differences were considered to be statistically significant when p-values were less than 0.05 (two-sided), resulting in 0.05/(number of contrasts performed) after Bonferroni correction. Statistical and graphical analyses were performed by using SPSS (version 16.0 for Windows, SPSS Inc., http://www.spss.com/) and Microsoft® Office Excel 2003 (Microsoft Corporation, http://office.microsoft.com/) software, respectively.
Results
The processing time for 42 samples was 40 min for the Reischl et al. method, 110 min for FTA® Elute and 5 h 30 min for the QIAamp® DNA Mini Kit. These processing times did not include the preparation of the solutions.
The sample input volumes were 90 μl for the Reischl et al. method, 40 μl for the FTA® Elute paper and 180 μl for the QIAamp® DNA Mini Kit. The output volumes for the samples were 100 μl for the Reischl et al. method, approximately 13.4 aliquots of 30 μl for the FTA® Elute paper and 100 μl for the QIAamp® DNA Mini Kit.
The concentration of the E. coli cell suspension was determined in duplicate at 1.09 × 107 cells 0.01 ml−1 (cumulating variation coefficient [CVC] = 10.52) and 9.51 × 106 cells 0.01 ml−1 (CVC = 9.76), with a mean of 1.02 × 109 cells ml−1. In addition, the concentration of the S. aureus cell suspension was determined in duplicate at 3.47 × 107 cells 0.01 ml−1 (CVC = 9.43) and 3.23 × 107 cells 0.01 ml−1 (CVC = 10.72), with a mean of 3.35 × 109 cells ml−1.
qrtPCR standard curves with E and R2 values that were determined for E. coli and S. aureus are presented in the Online Resources 1 and 2. The qrtPCR quantification limit was determined at 30 fg of the standard for E. coli and at 10 fg of the standard for S. aureus, respectively. Subsequently, when assuming a Ct equal to the mean Ct (+ 1 s.d.) of the qrtPCR quantification limit, extrapolated limits of quantification per extraction could be determined for E. coli to be 552 fg DNA for FTA® Elute and 137 fg DNA for the Reischl et al. method and QIAamp® DNA Mini Kit. For S. aureus, extrapolated limits of quantification per extraction were determined at 961 fg DNA for FTA® Elute and 238 fg DNA for the Reischl et al. method and QIAamp® DNA Mini Kit.
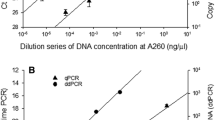
Figure 1a, b shows the median DNA levels measured per cell amount after extraction, their corresponding reference and the extrapolated limits of quantification for each DNA extraction method.
DNA levels extracted by three methods with varying amounts of cells. Median DNA levels as determined by qrtPCR resulting from quadruplicate extractions (n = 4) of different amounts of cells of (a) E. coli ATCC 11775T and (b) S. aureus subsp. aureus ATCC 25923. Extractions were performed by three different DNA extraction methods: FTA® Elute, the Reischl et al. method and QIAamp® DNA Mini Kit. The error bars indicate minimum and maximum values. Extrapolated quantification limits of the methods are represented by dotted lines: - - -, FTA® Elute; ─ ─ ─, Reischl et al. and QIAamp® DNA Mini Kit
The median efficiencies of DNA extraction of different cell amounts calculated for each DNA extraction method are presented in Table 1. Within each method, no significant differences were observed by the Kruskal–Wallis analysis amongst the efficiencies of DNA extraction of different cell amounts. Figure 2a, b gives an overview of the median efficiency of DNA extraction of the 5.3 × 105, 5.3 × 104 and 5.3 × 103 cell amounts for each method. p-values of p ≤ 0.017 were considered to be statistically significant for the performed three contrasts after Bonferroni correction. The experiments show that, for E. coli ATCC 11775T, the FTA® Elute paper had the highest median DNA extraction efficiency of 76.9%, followed by the Reischl et al. method with 43.7%. The QIAamp® DNA Mini Kit had the significantly lowest median DNA extraction efficiency of 7.7% as compared to FTA® Elute (P = 0.001) and the Reischl et al. method (P = 0.004). For S. aureus subsp. aureus ATCC 25923, the FTA® Elute paper and QIAamp® DNA Mini Kit performed with similar efficiencies of 108.9 and 97.7%, respectively. The efficiency of the Reischl et al. protocol was significantly lower with 9.0% as compared to the QIAamp® DNA Mini Kit (P = 0.001) and FTA® Elute paper (P = 0.001).
The level of PCR inhibition of the DNA extraction methods on the E. coli qrtPCR assay is presented in Fig. 3. The results show that, compared to added water to 1 ng of the qrtPCR standard DNA, both the added extraction controls of the Reischl et al. method and QIAamp® DNA Mini Kit caused a shift towards a higher Ct, indicating an inhibitory effect, in contrast to the extraction control of the FTA® Elute method. No inhibitory effects were noticeable on the S. aureus qrtPCR assay (data not shown).
Inhibition of the E. coli uidA qrtPCR assay. The mean levels of DNA were determined with qrtPCR by adding a template of 1 ng of the standard together with either water or an extraction control of one of the three following DNA extraction methods to triplicate reactions (n = 3): FTA® Elute, the Reischl et al. method and QIAamp® DNA Mini Kit. The dashed line represents part of the standard curve. (Ο) FTA® Elute, (□) QIAamp® DNA Mini Kit, (◊) Reischl, (Δ) Water
The applicability of the FTA® Elute method was examined by extracting DNA with this method from swab samples taken from the interior of the ISS. For S. aureus, a potential pathogen commonly found on the human skin, a location associated with human contact was positive with a mean of 4.50 ± 0.21 log fg DNA (~3.16 × 104 fg). Calculated from the chromosomal weight, this amount of DNA would correspond to ~1.05 × 104 S. aureus cells per sample. Sampling in the toilet area was positive for E. coli, with a mean of 4.50 ± 0.33 log fg DNA (~3.18 × 104 fg), as can be expected, since the potential pathogen E. coli is a common inhabitant of the human gut. This amount of DNA would correspond to ~5.58 × 103 E. coli cells per sample. In contrast, the mean S. aureus and E. coli DNA content from a location between wall panels was below the cut-off level.
Discussion
Molecular methods have been increasingly been used for the detection and quantification of hazardous microbes. Moreover, the need for a rapid and easy-to-use on-site molecular detection and quantification method is imperative. An equally important step in nucleic-acid-based quantification of bacteria is the part of sample processing preceding the actual measurement, as has previously been mentioned [3–5]. The results of this study confirm that DNA extraction is a key step in this process.
Several strategies can be used for the extraction of DNA from microbial samples, such as enzymatic, chemical or thermal lysis, mechanical disruption of the cell wall by beads or sonication, or a combination of the above [6, 9]. A disadvantage of enzymatic lysis is that commercially available enzymes can be contaminated with microbial DNA. Highly sensitive and specific nucleic-acid-based methods for the detection of bacteria necessitate the use of DNA extraction reagents that are free from contaminating bacterial nucleic acids. In addition, the use of enzymes often requires special conditions and preservation requirements, such as refrigeration and buffer storage. Chemical lysis often involves the usage of aggressive and toxic chemicals, which is less desirable for on-site detection where laboratory safety conditions are absent. A disadvantage for the application on-site to thermal lysis, mechanical disruption by beads or methods that use centrifugation is that the required equipment generally takes up a lot of weight and volume.
We selected to study three different DNA extraction methods to meet criteria for use on-site with qrtPCR on samples taken from solid surfaces, such as processing time, easiness of use, efficiency, range of bacterial species that it is effective for, chemical toxicity, and transport and storage requirements, which will be discussed below.
In our study, both the Reischl et al. protocol and FTA® Elute were, indeed, fast and easy to use by single-step isolation procedures and processing times of 40 and 110 min for 42 samples, respectively. DNA extraction methods with comparable processing times have been reported previously [3, 12–14].
Overall, FTA® Elute paper significantly had the highest efficiency to recover DNA from E. coli ATCC11755 and S. aureus subsp. aureus ATCC 25923 compared to the other two methods. As shown by the results, the measured amount of DNA in some cases exceeded that of the reference, which was estimated by the ‘gold standard’ of microscopic cell counting, resulting in percentages above 100%. An underestimation of the amount of chromosomal DNA by microscopic cell counting can easily occur due to factors such as cell clumping, cell division or cell lysis.
DNA extraction efficiency is limited by several factors. An important factor is cell concentration, which can be a delimiting factor for the quantification of natural samples where cell concentrations will vary. At low concentrations, method sensitivity will play a role and at high concentrations, overloading of the method can reduce DNA extraction efficiency. For FTA® Elute, problems can arise for certain harder-to-lyse species at concentrations above 107 cells ml−1 and overloading of the column of the QIAamp® DNA Mini Kit may lead to significantly lower yields than expected, according to the manufacturer’s instructions. A second factor is the type of bacterial species that is being subjected to the extraction. For instance, the constitution of the bacterial cell wall may impede cell lysis and the liberation of DNA from the cell. A third factor may be the physiological state of the cells that are subjected to cell lysis [7], in which context, it can be noted that the cells used in the present study were of liquid cultures in stationary phase. Another factor that may influence the outcome of quantification is the size of the DNA fragments generated by a specific method [6]. Long fragment sizes generated by the DNA extraction method may result in incomplete denaturation during the thermal cycling of qrtPCR, whilst, on the other hand, short fragment sizes may result in reduced amplification efficiency.
The QIAamp® DNA Mini Kit was developed and commonly used for a variety of bacterial species [15–24]. The FTA technology concept, including FTA® Elute, was also developed for use with a broad spectrum of bacterial species [25–27]. To our knowledge, the use of FTA® Elute for quantitative bacterial applications has not been assessed to date. The protocol by Reischl et al. was previously used for S. aureus; however, the main constituents and boiling procedure of the method have also been used for other species [9, 19, 28–33]. However, as confirmed by our results, it must be taken into account that inter-species and even intra-species differences in DNA extraction efficiencies will exist, influencing the outcome of microbial quantification [6, 8, 34]. Furthermore, in contrast to FTA® Elute, both the Reischl et al. and QIAamp® DNA Mini Kit method showed inhibitory effects on the E. coli qrtPCR assay but not on the S. aureus qrtPCR assay. Whilst the final volume of the FTA® Elute extract is four times more dilute than that of the other methods, the concentration of any inhibitory compounds it may possibly contain is too low to cause an inhibitory effect when used according to the manufacturer’s instructions. As such, inhibitory effects of a DNA extraction method can differ per PCR assay, as shown by the comparison of our results with those of other authors [8, 16]. PCR inhibition reduces sensitivity, thereby negatively influencing the outcome of quantitative measurements. As a result, different rates of PCR inhibition on separate PCR assays will complicate the quantification of microbial samples with a complex composition. Moreover, these observations should also be carefully considered in the choice of inhibition controls [3]. In addition to PCR inhibition generated by the DNA extraction method itself, as described above, PCR inhibition needs to be assessed depending on the specimen type. In this study, we sought to select a DNA extraction method suitable for the processing of samples taken from solid surfaces in various surroundings. However, a wide variety of possible PCR inhibiting compounds or, most often, none at all can be emphasised to be present on such surfaces, leaving any prediction on the PCR inhibition generated by this particular specimen type speculative.
The least toxic method appears to be FTA® Elute, which, apart from the proprietary chemicals on the paper, only requires the use of water. The protocol developed by Reischl et al. utilises the detergents Tween 20 and Triton X-100, whilst the QIAamp® DNA Mini Kit involves the use of irritating substances.
The protocol according to Reischl et al. and the FTA® Elute method both occupy little volume and weight. The Reischl et al. protocol only requires one lysis buffer, thermal heating and centrifugation. FTA® Elute consists of small paper sample cards and, in addition, requires water, a small puncher, thermal heating and centrifugation. Additional transport and storage advantages for the FTA® Elute method are room temperature sample storage, protection of DNA against radiation, UV damage and enzymatic breakdown, and the immediate deactivation of potentially hazardous microbes, according to the manufacturer. The QIAamp® DNA Mini Kit makes use of several solutions, spin-columns, centrifugation and thermal heating steps and, as such, occupies more volume and weight than the other methods.
Finally, the applicability of the FTA® Elute method for the analysis of environmental samples was demonstrated with swab samples taken from the interior of the ISS. The bacterial levels that correspond to the amount of DNA measured in these samples are also in agreement with hygiene measures taken aboard the ISS aimed to keep bacterial levels below the bacterial acceptability limit of 104 colony forming units per 100 cm2 [35].
The importance of the DNA extraction step in nucleic-acid-based molecular detection and quantification methodology for bacteria and the importance of the assessment of its efficiency is evident, as is the case in fungal diagnostics [36]. Following the results of this study, FTA® Elute paper would be our method of choice for use on-site with samples taken from solid surfaces, based on the processing time per sample, easiness of use, DNA extraction efficiency, broad range of species effectiveness, chemical toxicity, and transport and storage advantages.
References
Zengler K, Rajilic-Stojanovic M, De Vos WM, Zoetendal EG, Sloan WT, Quince C, Curtis TP, Schmidt SK, Wilson KL, Meyer AF, Schadt CW, Porter TM, Moncalvo JM, Caron DA, Gast RJ, Fierer N, Rosselló-Mora R, López-López A, Fuhrman JA, Nelson KE, Janssen PH, Ciche TA, Kartal B, Strous M, Avery SV, Desikan R, Tetard L, Passian A, Datar RH, Thundat T, Keller M, Schadt CW, Palumbo AV, Smith HO, Glass JI, Hutchison CA 3rd, Venter JC (2008) Accessing uncultivated microorganisms: from the environment to organisms and genomes and back. ASM Press, Washington, DC
van Tongeren SP, Krooneman J, Raangs GC, Welling GW, Harmsen HJM (2007) Microbial detection and monitoring in advanced life support systems like the International Space Station. Microgravity Sci Technol 19:45–48
Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR 3rd, Smith TF (2006) Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev 19:165–256
Bastien P, Procop GW, Reischl U (2008) Quantitative real-time PCR is not more sensitive than “conventional” PCR. J Clin Microbiol 46:1897–1900
Cook KL, Britt JS (2007) Optimization of methods for detecting Mycobacterium avium subsp. paratuberculosis in environmental samples using quantitative, real-time PCR. J Microbiol Methods 69:154–160
Rantakokko-Jalava K, Jalava J (2002) Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J Clin Microbiol 40:4211–4217
Ludwig W, Schleifer KH (2000) How quantitative is quantitative PCR with respect to cell counts? Syst Appl Microbiol 23:556–562
Coyne SR, Craw PD, Norwood DA, Ulrich MP (2004) Comparative analysis of the Schleicher and Schuell IsoCode Stix DNA isolation device and the Qiagen QIAamp DNA Mini Kit. J Clin Microbiol 42:4859–4862
Reischl U, Linde HJ, Metz M, Leppmeier B, Lehn N (2000) Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J Clin Microbiol 38:2429–2433
Harmsen HJM, Raangs GC, He T, Degener JE, Welling GW (2002) Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol 68:2982–2990
Brasher CW, DePaola A, Jones DD, Bej AK (1998) Detection of microbial pathogens in shellfish with multiplex PCR. Curr Microbiol 37:101–107
Hogg GM, McKenna JP, Ong G (2008) Rapid detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive BacT/Alert® blood culture bottles using real-time polymerase chain reaction: evaluation and comparison of 4 DNA extraction methods. Diagn Microbiol Infect Dis 61:446–452
Cremonesi P, Castiglioni B, Malferrari G, Biunno I, Vimercati C, Moroni P, Morandi S, Luzzana M (2006) Technical note: improved method for rapid DNA extraction of mastitis pathogens directly from milk. J Dairy Sci 89:163–169
Cenci-Goga BT, Crotti S, Costarelli S, Rondini C, Karama M, Bennett P (2004) Detection of tet(M) gene from raw milk by rapid DNA extraction followed by a two-step PCR with nested primers. J Food Prot 67:2833–2838
Gullsby K, Hallander HO, Bondeson K (2007) Performance of Bordetella pertussis IS481 real-time PCR in a vaccine trial setting. APMIS 115:1370–1375
Aldous WK, Pounder JI, Cloud JL, Woods GL (2005) Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J Clin Microbiol 43:2471–2473
Lee SH, Jung BY, Rayamahji N, Lee HS, Jeon WJ, Choi KS, Kweon CH, Yoo HS (2009) A multiplex real-time PCR for differential detection and quantification of Salmonella spp., Salmonella enterica serovar Typhimurium and Enteritidis in meats. J Vet Sci 10:43–51
Luan C, Levin RE (2008) Use of activated carbon coated with bentonite for increasing the sensitivity of pcr detection of Escherichia coli O157:H7 in Canadian oyster (Crassostrea gigas) tissue. J Microbiol Methods 72:67–72
Wilson DA, Yen-Lieberman B, Reischl U, Gordon SM, Procop GW (2003) Detection of Legionella pneumophila by real-time PCR for the mip gene. J Clin Microbiol 41:3327–3330
Wilson D, Yen-Lieberman B, Reischl U, Warshawsky I, Procop GW (2004) Comparison of five methods for extraction of Legionella pneumophila from respiratory specimens. J Clin Microbiol 42:5913–5916
Peters RPH, van Agtmael MA, Gierveld S, Danner SA, Groeneveld ABJ, Vandenbroucke-Grauls CMJE, Savelkoul PHM (2007) Quantitative detection of Staphylococcus aureus and Enterococcus faecalis DNA in blood to diagnose bacteremia in patients in the intensive care unit. J Clin Microbiol 45:3641–3646
Morio F, Corvec S, Caroff N, Le Gallou F, Drugeon H, Reynaud A (2008) Real-time PCR assay for the detection and quantification of Legionella pneumophila in environmental water samples: utility for daily practice. Int J Hyg Environ Health 211:403–411
Simms I, Eastick K, Mallinson H, Thomas K, Gokhale R, Hay P, Herring A, Rogers PA (2003) Associations between Mycoplasma genitalium, Chlamydia trachomatis, and pelvic inflammatory disease. Sex Transm Infect 79:154–156
Dauphin LA, Stephens KW, Eufinger SC, Bowen MD (2010) Comparison of five commercial DNA extraction kits for the recovery of Yersinia pestis DNA from bacterial suspensions and spiked environmental samples. J Appl Microbiol 108:163–172
Rajendram D, Ayenza R, Holder FM, Moran B, Long T, Shah HN (2006) Long-term storage and safe retrieval of DNA from microorganisms for molecular analysis using FTA matrix cards. J Microbiol Methods 67:582–592
Jaravata CV, Smith WL, Rensen GJ, Ruzante JM, Cullor JS (2006) Detection of Mycobacterium avium subsp. paratuberculosis in bovine manure using Whatman FTA card technology and Lightcycler real-time PCR. Foodborne Pathog Dis 3:212–215
Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, Matherly LH, Majumdar APN, Kato I (2008) Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods 72:124–132
Reischl U, Pulz M, Ehret W, Wolf H (1994) PCR-based detection of mycobacteria in sputum samples using a simple and reliable DNA extraction protocol. Biotechniques 17:844–845
Miller MB, Meyer H, Rogers E, Gilligan PH (2005) Comparison of conventional susceptibility testing, penicillin-binding protein 2a latex agglutination testing, and mecA real-time PCR for detection of oxacillin resistance in Staphylococcus aureus and coagulase-negative Staphylococcus. J Clin Microbiol 43:3450–3452
Sakai H, Procop GW, Kobayashi N, Togawa D, Wilson DA, Borden L, Krebs V, Bauer TW (2004) Simultaneous detection of Staphylococcus aureus and coagulase-negative staphylococci in positive blood cultures by real-time PCR with two fluorescence resonance energy transfer probe sets. J Clin Microbiol 42:5739–5744
Shrestha NK, Tuohy MJ, Padmanabhan RA, Hall GS, Procop GW (2005) Evaluation of the LightCycler Staphylococcus MGRADE kits on positive blood cultures that contained gram-positive cocci in clusters. J Clin Microbiol 43:6144–6146
Shrestha NK, Tuohy MJ, Hall GS, Reischl U, Gordon SM, Procop GW (2003) Detection and differentiation of Mycobacterium tuberculosis and nontuberculous mycobacterial isolates by real-time PCR. J Clin Microbiol 41:5121–5126
Shrestha NK, Tuohy MJ, Hall GS, Isada CM, Procop GW (2002) Rapid identification of Staphylococcus aureus and the mecA gene from BacT/ALERT blood culture bottles by using the LightCycler system. J Clin Microbiol 40:2659–2661
Zucol F, Ammann RA, Berger C, Aebi C, Altwegg M, Niggli FK, Nadal D (2006) Real-time quantitative broad-range PCR assay for detection of the 16S rRNA gene followed by sequencing for species identification. J Clin Microbiol 44:2750–2759
Pierson DL (2001) Microbial contamination of spacecraft. Gravit Space Biol Bull 14:1–6
Fredricks DN, Smith C, Meier A (2005) Comparison of six DNA extraction methods for recovery of fungal DNA as assessed by quantitative PCR. J Clin Microbiol 43:5122–5128
Acknowledgements
We would especially like to thank A. M. Borries, S. van der Schaaf and G. C. Raangs for their technical contributions to this project. We thank W. J. Sluiter for the statistical advice and G. W. Welling for critically reading the manuscript. None of the authors had any degree of commercial or potential dual interest with the manufacturers mentioned in this study. This project was supported by the European Space Agency (MAP project number AO-LS-99-MAP-LSS-018 ‘Biofilms’).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
qrtPCR standard curve with E and R2 values of E. coli (n = 6). (GIF 61 KB)
Online Resource 2
qrtPCR standard curve with E and R2 values of S. aureus (n = 9). (GIF 60 KB)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van Tongeren, S.P., Degener, J.E. & Harmsen, H.J.M. Comparison of three rapid and easy bacterial DNA extraction methods for use with quantitative real-time PCR. Eur J Clin Microbiol Infect Dis 30, 1053–1061 (2011). https://doi.org/10.1007/s10096-011-1191-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-011-1191-4