Abstract
From January 2004 to December 2005, a subset of stool specimens (n = 189) from patients who attended an urban hospital in Bangladesh, in which no pathogen was detected, was tested for the presence of noroviruses by conventional reverse transcription–polymerase chain reaction (RT-PCR). Norovirus RNA was detected in 37 samples (19.6%) in the no-pathogen-detected samples and the estimated overall norovirus detection rate was 8.5%. Diarrhea was generally moderate in the norovirus-infected patients and vomiting was the most common feature among them. Genetic analysis indicated that the GII genogroup was the most predominant norovirus strain (82.4%). The GI strain was found in 17.6% of samples and no cases of GIV were detected. This study indicates that a remarkable proportion of the diarrhea patients is hospitalized due to norovirus infection. Therefore, routine diagnosis of this virus in hospitalized patients is required. Since our study was based on hospitalized patients, community surveillance would be helpful to estimate the true burden of the virus in the country. The data regarding the genetic information of the circulating norovirus strains would be very useful for the norovirus vaccine development programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human noroviruses (NoVs), members of the family Caliciviridae, are now recognized as the most common cause of outbreaks of nonbacterial gastroenteritis worldwide [1, 2]. NoV infections are especially associated with the ingestion of contaminated water [3–6], food [7, 8], and oysters [9, 10]. Since a low infectious dose is enough to develop illness in humans, the virus is highly contagious [11, 12]. It is estimated that noroviruses cause about 900,000 episodes of gastroenteritis that require a clinic visit and 64,000 hospitalizations among children <5 years of age residing in high-income countries each year. In developing countries, noroviruses are estimated to cause 200,000 child deaths each year [13].
NoVs have been detected in persons of all ages, but more frequently in children <5 years of age [13]. Clinical features associated with noroviruses include nausea, vomiting, abdominal cramps, myalgias, headache, fever, chills, sore throat, and non-bloody diarrhea [14–16]. Although NoV transmission occurs year-round in most parts of the world, generally, a cold weather peak was found in The Netherlands, England and Wales, Japan, the US, Australia, Canada, and Denmark [17, 18].
The norovirus genome is positive-sense, single-stranded RNA, approximately 7.7 kb in length [19]. There exists considerable diversity among norovirus strains circulating in different geographical locations and time periods. Five genogroups (GI through GV) of NoVs have been assigned from the molecular characterization of complete capsid gene sequences, which are further subdivided into 27 genotypes [20–22]. Human disease has been associated with GI, GII, and GIV, of which genetic cluster GII.4 are the most predominant strains [23–27]. Recently, a new GII.4 variant with mutation in the polymerase gene circulating in Europe was responsible for 50% of outbreaks in the United Kingdom [14, 28]. Similarly, a novel recombinant GII.3 strain was identified in 2006 in 44% of the clinical samples in Japan [29].
Reverse transcription–polymerase chain reaction (RT-PCR) is currently the most widely used technique for the detection of norovirus from stool, water, and food [30–32]. Several primer pairs have been described that were deduced from highly conserved polymerase gene [33–35]. However, none of the reported conventional RT-PCR assays were able to detect all strains. Sequence analysis of the polymerase region of a wide range of virus strains indicated that this region was also variable, with nucleotide identity as low as 53% between strains of different genogroups and 60–64% within genogroups [36]. In recent years, real-time RT-PCR methodology has emerged as a potentially important diagnostic procedure [37, 38].
Diarrhea is an important public health concern in developing countries, including Bangladesh. Over 100,000 diarrhea patients are admitted at the Dhaka hospital of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B) each year, and stool samples are investigated in the hospital surveillance system for the presence of different enteric pathogens (Salmonella typhi, non-typhi Salmonella, Shigella dysenteriae, Shigella flexneri, Shigella boydii, Shigella sonnei, Vibrio cholerae O139, Vibrio cholerae O1, and group A rotavirus). It was observed that a huge proportion of samples was left undiagnosed each year. It is possible that other diarrhea-causing agents, such as norovirus, adenovirus, sapovirus, astrovirus, group B rotavirus, group C rotavirus, Aichi virus, etc., which were not included in the hospital surveillance system could have been involved in causing diarrhea hospitalization. To investigate the burden of these unknown pathogens, we aimed to detect noroviruses in these undiagnosed specimens. We also characterized the Bangladeshi NoV strains by sequencing their polymerase genes and compared them with other globally circulating strains to reveal their genetic relationships.
Methods
RT-PCR
Viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. A quantity of 140 µl of stool specimen was taken for extraction and final elution was done with 50 µl RNase-free water. RT-PCR was carried out using the OneStep RT-PCR Kit (Qiagen, Hilden, Germany). Forward primers Mon 431 (5’-TGG ACI AGR GGI CCY AAY-3’) and Mon 432 (5’-TGG ACI CGY GGI CCY AAY-3’) and reverse primers Mon 433 (5’-GAA YCT CAT CCA YCT GAA-3’) and Mon 434 (5’-GAA SCG CAT CCA RCG GAA-3’) were used to amplify 213-nucleotide-long segments from the ORF1 of the norovirus genome [33]. The reaction was carried out with an initial reverse transcription step at 50°C for 30 min with an activation step at 95°C for 15 min, followed by 40 cycles of amplification (30 s at 94°C, 30 s at 52°C, 30 s at 72°C) and a final extension of 10 min at 72°C in a thermal cycler. PCR products were run on a 1.5% ethidium bromide-stained agarose gel and visualized under UV light.
Nucleotide sequencing
The PCR products were purified with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced using the dideoxynucleotide chain termination method with the ABI PRISM® BigDye Terminator Cycle Sequencing Reaction Kit (Applied Biosystems, Foster City, CA) on an automated sequencer. The same forward primers which were used in the PCR amplification were used as sequencing primers.
DNA and protein sequence analysis
The chromatogram sequencing files were inspected using Chromas 2.23 (Technelysium, Queensland, Australia) and consensus sequences were prepared using SeqMan II (DNASTAR, Madison, WI). Nucleotide and amino acid sequence similarity searches were performed using the National Center for Biotechnology Information (NCBI; National Institutes of Health, Bethesda, MD) BLAST (basic local alignment search tool) server on the GenBank database, release 173.0 [39]. Multiple sequence alignments were calculated using ClustalX 1.81 [40]. Sequences were manually edited in the GeneDoc version 2.6.002 alignment editor [41]. Phylogenetic analyses were conducted using the MEGA version 4.1 software package [42]. The dendrograms were constructed using the neighbor-joining method.
Nucleotide sequence accession numbers
The nucleotide sequences reported in this paper were submitted to GenBank using National Center for Biotechnology Information (NCBI; Bethesda, MD) Sequin version 9.2 and were assigned under accession numbers GU370930–GU370937.
Data analysis
The data were analyzed using SPSS for Windows release 11.5.1 (SPSS Inc., Chicago, IL).
Results
Detection of norovirus
From January 2004 to December 2005, a total of 4,407 diarrhea patients attended the Dhaka hospital of the ICDDR,B. The hospital surveillance system tested 2% samples for several pathogens (i.e., group A rotaviruses, Salmonella typhi, non-typhi Salmonella, Shigella dysenteriae, Shigella flexneri, Shigella boydii, Shigella sonnei, Vibrio cholerae O139, and Vibrio cholerae O1). Any of these pathogens were detected in 2,468 (56%) samples and the remaining 1,939 (44%) were undiagnosed. From these undiagnosed samples, every tenth stool specimen (n = 189) was selected for the presence of NoV by conventional RT-PCR. Norovirus RNA was detected in 37 samples (19.6%), of which 18 were detected in 2004 (n = 100) and 19 in 2005 (n = 89).
Seasonality
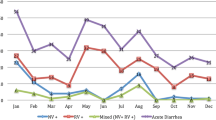
The virus was isolated throughout the year and no clear seasonality was observed for NoV infection, as depicted in Fig. 1. However, comparatively high numbers were detected in August–September (rainy season) and December (winter).
Age distribution
NoV was detected in both children and adult patients, with the age ranging from 1.8 months to 50 years. The median age of the norovirus-positive patients was 2 years and the mean age was 14 years. Figure 2 shows the age distribution of the NoV-infected patients. Remarkably, children less than 2 years of age (49% of the NoV-positive patients) and elderly people more than 18 years of age (43% of the NoV-positive patients) were more likely to be infected by NoV. Only three patients were between 2 and 18 years old.
Clinical features
Among the NoV-positive patients, 62% were male. Clinical features of the norovirus-infected patients are shown in Table 1. The illness caused by norovirus infection was mild and was mainly characterized by diarrhea and vomiting. Vomiting was the most common feature and was observed in 83.8% of patients. Fever (19.1%) and abdominal cramp (37.8%) were not frequent in these patients. Severe dehydration caused by norovirus gastroenteritis was found in about 20% of patients and most of them were in the elderly age group (6 out of 7 severely ill patients). Diarrhea was generally moderate; 73% of patients had diarrhea for less than 3 days and 35% of patients had more than ten stools over a 24-h period. Since NoV caused mild dehydration in our study, most of the patients were treated by oral rehydration solution (ORS) (70.3%) only. For severe cases, intravenous fluid (IV) was given additionally to the rest of the patients.
Genetic characterization
The RNA samples which produced sufficient PCR-amplified products (n = 17) were sequenced using the dideoxynucleotide chain termination method. Mon 431 and Mon 432 primers produced at least 100 nucleotide bases from the 5’-end of the polymerase gene. Nucleotide similarity searches were done using the NCBI BLAST server on GenBank database release 130.0 [39]. Similarity searches indicated that three (17.6%) were NoV genogroup GI and 14 (82.4%) were genogroup GII (Table 2). Among the GI strains, two were most similar to genotype GI.1 and one was GI.3. Among the GII strains, four were GII.4 (28.6%) and nine were GII.7 (64.3%). One sample was found to be mixed with GI and GII genogroups.
BLAST analysis indicated that the GI Bangladeshi strains Dhaka54 and Dhaka67 were most similar to Indian V16/06/IND (94% nucleotide and 100% amino acid identity), Russian 6836/Chelyabinsk/RUS (96% nucleotide and 100% amino acid identity), and GI.1 reference US NoV strain West Chester/2001/USA (96% nucleotide and 100% amino acid identity). Another GI isolate, Dhaka23, had 98% nucleotide and 100% amino acid identity with Russian strain 11227/NizhnyNovgorod/RUS. Nucleotide identities with other strains were low (less than 85%). An Indian strain Kolkata/L8775/2006/IND had 80 and 98% identities with Dhaka23 based on nucleotides and amino acids, respectively.
The GII strains isolated in Bangladesh were divided into two genotypes, GII.4 and GII.7. Based on amino acid sequences, isolates Dhaka58 and Dhaka100 were most similar to GII.4 Indian strain V1699/07/IND, as well as strains from all over the world (100% identity). The most similar strain based on nucleotide identity was Canadian Manitoba/4205/2003/ CAN. On the other hand, Dhaka55 and Dhaka97 were most similar to GII.7 East Asian norovirus strains. Absolute identity on amino acid sequences was identified with Japanese Saitama U25, Chinese Shanxi/50106/2006, and Thai strain Mc17/2002/Th. Isolate Dhaka42 was placed a little farther from them and was most similar to Brazilian strain 5037/2001/Bra (99% nucleotide and 100% amino acid identity).
Three GI sequences and five GII sequences were included in the phylogenetic trees, along with globally circulating NoV strains (Figs. 3 and 4). GI Bangladeshi strains belonged to two different clusters. Dhaka54 and Dhaka67 were closely related to a US strain WestChester of GI.1 lineage. The third Bangladeshi GI strain, Dhaka23, did not cluster closely with any genotype, although the mostly related genotype was GI.3. Bangladeshi GII NoVs clustered with two different genotypes. Dhaka58 and Dhaka100 were closely related to the GII.4 cluster and Dhaka97 and Dhaka55 were closely related to the GII.7 cluster.
Phylogenetic tree based on partial nucleotide sequences of the polymerase gene of norovirus genogroup GI. The tree was constructed by the neighbor-joining method. The numbers adjacent to the nodes represent the percentages of bootstrap support (of 1,000 replicates) for the clusters to the right of the nodes. Bootstrap values lower than 75% are not shown. Bangladeshi strains are in bold. BGD, Bangladesh; USA, United States of America; JPN, Japan; CAN, Canada; NZL, New Zealand; BRA, Brazil; GBR, Great Britain; DEU, Germany; DJI, Djibouti; FRA, France. The GenBank accession numbers were: West Chester (AY502016), Saitama T23cGI (AB112094), Saitama KU19dGI (AB058528), Saitama T24cGI (AB112098), Saitama T2GI (AB112103), Little Rock316 (AF414405), NoV69 (EF078281), DjiboutiVdG50 (EF190919), ICB1969C2 (DQ386978), Wyoming (AY210317), PontdeRoide682 (EF529736), Berlin/92617.04-3-BA2 (DQ340078), Baltimore/277 (AF414404), Saitama T24dGI (AB112099), NoV748 (EF078287), BCCDC04003 (DQ452547), Wisconsin (AY502008), Queen’s Arms/Leeds (AJ313030), Appalachicola Bay/318 (AF414406), Cardrona (EF527258), Berlin/92674.04-5-BA2 (DQ340080), ICB1918 (DQ386948), New Orleans/266 (AF414402), Berlin/90068.04-8-BA4 (DQ340083), Steinbach/EG (AF473567), and an outgroup sequence from a GII norovirus Saitama T24eGII (AB112306) is included
Phylogenetic tree based on partial nucleotide sequences of the polymerase gene of norovirus genogroup GII. The tree was constructed by the neighbor-joining method. The numbers adjacent to the nodes represent the percentages of bootstrap support (of 1,000 replicates) for the clusters to the right of the nodes. Bootstrap values lower than 75% are not shown. Bangladeshi strains are in bold. AUS, Australia; NLD, The Netherlands; IRE, Ireland; VIE, Vietnam; ITA, Italy; HUN, Hungary; CHN, China. Reference strains of GII genotypes were collected from GenBank and the accession numbers of the reference strains are as follows: Hawaii (U07611), PC301 (AF414421), MOH99 (AF397156), NO306 (AF414422), SU16 (AB039778), SU25 (AB039780), SW918 (AB074893), SU1 (AB039775), HLL314 (AF414420), Tiffin (AY502010), CSE1 (AY502009), Manitoba4205 (DQ463420), OCS980730 (AB089871), Sydney348-97O (DQ078829), Gwynedd273 (AF414409), 691 (AF493210), Terneuzen70 (EF126964), BCCDC04007 (DQ452543), ICB2162 (DQ386959), NoV809 (EF078293), Rhyl440 (DQ822040), Carlow (DQ415279), Kunming146 (DQ304651), NV/Osaka/F140 (AB258331), Minato/N1-14 (AB233474), Burwash Landing331 (AF414425), Dresden174-pUS-NorII (AY741811), Miami81 (AF414416), and Vietnam 026 (AF504671), with an outgroup GI Baltimore/277 (AF414404)
Discussion
From 1993 to 2004, among diarrhea patients less than 5 years of age who attended the Dhaka hospital of the ICDDR,B, n = 18,544 were tested for several pathogens. Rotavirus constituted 33%, bacterial agents, including V. cholerae, Shigella, and Salmonella, 21%, and parasites 2% [43]. Remarkably, a major proportion (44% of the samples tested) was undiagnosed. Our results show that NoVs constituted 19.6% of these undiagnosed samples and are a common diarrhea-causing agent which necessitates hospitalization. Overall, the prevalence of NoV in all pathogen-positive and -negative samples (n = 440) was 8.4%. The overall NoV detection rate of 8.4% in our study is similar to other studies which report NoV prevalence in the range 6–19% [44–52]. The detection rate in our study, however, might be underestimated because the primer set that we used might not be able to detect all of the different strains. It is noteworthy that designing one set of primers to detect all NoV strains with equal efficiency is difficult because the mutation rate in the NoV genome is very high [53, 54].
Norovirus was found in both children and adults in our study. Similarly, reports from all over the world also indicated that NoVs could infect both children and adults [55–58]. Clinical features of the NoV-infected patients were characterized by diarrhea, abdominal cramp, vomiting, and fever. Although hospitalized, the illness of most of our NoV-infected patients was not severe, which was supported by the previous investigations [59–61]. Treatments of the patients were carried out by ORS in most cases and a few severe patients were treated with IV.
It is established that GI, GII, and GIV are the major norovirus genogroups isolated worldwide causing human gastroenteritis [20]. In the present study, we also identified norovirus genogroups GII and GI, although GII strains were predominant over GI strains. Genogroup GI was present in only small numbers in our study period, which is also similar to other studies conducted during the same period [11, 24, 54, 62–64]. It is interesting to note that a previous study conducted in Bangladeshi children less than 38 months of age found only the GII.4 genotype [59]. The GII.4 strains identified in the present study were also found in infants and children (aged 4 to 12 months). No GIV strain was detected in our study, although this genogroup has been circulating in humans worldwide [20, 65].
One of the goals of this study was to compare Bangladeshi strains with other noroviruses isolated all over the world to reveal their origin and genetic relationships. Phylogenetic analysis indicated that the Bangladeshi strains did not cluster with strains from a particular region; instead, strains from different countries clustered in the same branch. This indicates that the global dispersion of the viruses occurred between different geographical locations. GI strain Dhaka23 was an interesting strain, since it was placed in a separate branch distantly related to other GI genotypes (Fig. 3). Since we analyzed a small portion of the polymerase gene, complete genome sequence analysis would be required for detailed characterization of this strain.
The study had several limitations. The sample size was small and was collected over a 24-month period. Longer, longitudinal studies are required to address issues such as norovirus seasonality and genetic variability and to monitor the spread and persistence of the various genotypes circulating in Bangladesh. In addition, we sequenced a small proportion of the norovirus genes which might have an impact on genotyping of the strains described in this paper. The use of short conserved sequences, although successful for the diagnosis of norovirus infection, should be used with caution for classification and phylogenetic analyses. Therefore, an expanded genetic study is required to genotype the strains accurately, as well as to detect new and novel strains, and further analysis with full genome sequences might be more helpful. Since the study included only hospitalized patients, community-based studies are required in order to investigate the true burden of the disease caused by this pathogen. In conclusion, this study provides information for the future epidemiological studies of noroviruses, which is required for the control and prevention of diarrheal diseases, as well as norovirus vaccine development programs.
References
Green KY (2007) Caliciviridae: the noroviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (eds) Fields virology. Lippincott Williams & Wilkins, Philadelphia, pp 949–979
Hutson AM, Atmar RL, Estes MK (2004) Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol 12(6):279–287
Hernandez-Morga J, Leon-Felix J, Peraza-Garay F, Gil-Salas BG, Chaidez C (2009) Detection and characterization of hepatitis A virus and Norovirus in estuarine water samples using ultrafiltration—RT-PCR integrated methods. J Appl Microbiol 106(5):1579–1590
Nygård K, Torvén M, Ancker C, Knauth SB, Hedlund KO, Giesecke J, Andersson Y, Svensson L (2003) Emerging genotype (GGIIb) of norovirus in drinking water, Sweden. Emerg Infect Dis 9(12):1548–1552
Miettinen IT, Zacheus O, von Bonsdorff CH, Vartiainen T (2001) Waterborne epidemics in Finland in 1998–1999. Water Sci Technol 43(12):67–71
Ramia S (1985) Transmission of viral infections by the water route: implications for developing countries. Rev Infect Dis 7(2):180–188
Cliver DO (1997) Virus transmission via food. World Health Stat Q 50(1–2):90–101
Parashar UD, Monroe SS (2001) “Norwalk-like viruses” as a cause of foodborne disease outbreaks. Rev Med Virol 11(4):243–252
Lowther JA, Henshilwood K, Lees DN (2008) Determination of norovirus contamination in oysters from two commercial harvesting areas over an extended period, using semiquantitative real-time reverse transcription PCR. J Food Prot 71(7):1427–1433
Dowell SF, Groves C, Kirkland KB, Cicirello HG, Ando T, Jin Q, Gentsch JR, Monroe SS, Humphrey CD, Slemp C, Dwyer DM, Meriwether RA, Glass RI (1995) A multistate outbreak of oyster-associated gastroenteritis: implications for interstate tracing of contaminated shellfish. J Infect Dis 171(6):1497–1503
Bull RA, Tu ET, McIver CJ, Rawlinson WD, White PA (2006) Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J Clin Microbiol 44(2):327–333
Beuret C, Kohler D, Baumgartner A, Lüthi TM (2002) Norwalk-like virus sequences in mineral waters: one-year monitoring of three brands. Appl Environ Microbiol 68(4):1925–1931
Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinjé J, Parashar UD (2008) Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis 14(8):1224–1231
Lopman B, Vennema H, Kohli E, Pothier P, Sanchez A, Negredo A, Buesa J, Schreier E, Reacher M, Brown D, Gray J, Iturriza M, Gallimore C, Bottiger B, Hedlund KO, Torvén M, von Bonsdorff CH, Maunula L, Poljsak-Prijatelj M, Zimsek J, Reuter G, Szücs G, Melegh B, Svennson L, van Duijnhoven Y, Koopmans M (2004) Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363(9410):682–688
Kapikian AZ (1996) Overview of viral gastroenteritis. Arch Virol Suppl 12:7–19
Kapikian AZ (2000) The discovery of the 27-nm Norwalk virus: an historic perspective. J Infect Dis 181(Suppl 2):S295–S302
Lopman BA, Reacher MH, Van Duijnhoven Y, Hanon FX, Brown D, Koopmans M (2003) Viral gastroenteritis outbreaks in Europe, 1995–2000. Emerg Infect Dis 9(1):90–96
Mounts AW, Ando T, Koopmans M, Bresee JS, Noel J, Glass RI (2000) Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J Infect Dis 181(Suppl 2):S284–S287
Jiang X, Wang M, Wang K, Estes MK (1993) Sequence and genomic organization of Norwalk virus. Virology 195(1):51–61
Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS (2006) Norovirus classification and proposed strain nomenclature. Virology 346(2):312–323
Fankhauser RL, Monroe SS, Noel JS, Humphrey CD, Bresee JS, Parashar UD, Ando T, Glass RI (2002) Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J Infect Dis 186(1):1–7
Vinjé J, Koopmans MP (2000) Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J Clin Microbiol 38(7):2595–2601
Cubitt WD, Green KY, Payment P (1998) Prevalence of antibodies to the Hawaii strain of human calicivirus as measured by a recombinant protein based immunoassay. J Med Virol 54(2):135–139
Ike AC, Brockmann SO, Hartelt K, Marschang RE, Contzen M, Oehme RM (2006) Molecular epidemiology of norovirus in outbreaks of gastroenteritis in southwest Germany from 2001 to 2004. J Clin Microbiol 44(4):1262–1267
Dimitrov DH, Dashti SA, Ball JM, Bishbishi E, Alsaeid K, Jiang X, Estes MK (1997) Prevalence of antibodies to human caliciviruses (HuCVs) in Kuwait established by ELISA using baculovirus-expressed capsid antigens representing two genogroups of HuCVs. J Med Virol 51(2):115–118
Jiang X, Wilton N, Zhong WM, Farkas T, Huang PW, Barrett E, Guerrero M, Ruiz-Palacios G, Green KY, Green J, Hale AD, Estes MK, Pickering LK, Matson DO (2000) Diagnosis of human caliciviruses by use of enzyme immunoassays. J Infect Dis 181(Suppl 2):S349–S359
O’Ryan ML, Víal PA, Mamani N, Jiang X, Estes MK, Ferrecio C, Lakkis H, Matson DO (1998) Seroprevalence of Norwalk virus and Mexico virus in Chilean individuals: assessment of independent risk factors for antibody acquisition. Clin Infect Dis 27(4):789–795
Kirkwood C (2004) Viral gastroenteritis in Europe: a new norovirus variant? Lancet 363(9410):671–672
Phan TG, Kuroiwa T, Kaneshi K, Ueda Y, Nakaya S, Nishimura S, Yamamoto A, Sugita K, Nishimura T, Yagyu F, Okitsu S, Müller WE, Maneekarn N, Ushijima H (2006) Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J Med Virol 78(7):971–978
De Leon R, Matsui SM, Baric RS, Herrmann JE, Blacklow NR, Greenberg HB, Sobsey MD (1992) Detection of Norwalk virus in stool specimens by reverse transcriptase-polymerase chain reaction and nonradioactive oligoprobes. J Clin Microbiol 30(12):3151–3157
Schwab KJ, De Leon R, Sobsey MD (1996) Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl Environ Microbiol 62(6):2086–2094
Schwab KJ, Neill FH, Fankhauser RL, Daniels NA, Monroe SS, Bergmire-Sweat DA, Estes MK, Atmar RL (2000) Development of methods to detect “Norwalk-like viruses” (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak. Appl Environ Microbiol 66(1):213–218
Chatterjee NK, Moore DW, Monroe SS, Glass RI, Cambridge MJ, Kondracki SF, Morse DL (2004) Molecular epidemiology of outbreaks of viral gastroenteritis in New York State, 1998–1999. Clin Infect Dis 38(Suppl 3):S303–S310
Ando T, Noel JS, Fankhauser RL (2000) Genetic classification of “Norwalk-like viruses”. J Infect Dis 181(Suppl 2):S336–S348
Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO (1999) Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J Virol Methods 83(1–2):145–154
Vinjé J, Green J, Lewis DC, Gallimore CI, Brown DW, Koopmans MP (2000) Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses”. Arch Virol 145(2):223–241
Marshall JA, Bruggink LD (2006) Laboratory diagnosis of norovirus. Clin Lab 52(11–12):571–581
Trujillo AA, McCaustland KA, Zheng DP, Hadley LA, Vaughn G, Adams SM, Ando T, Glass RI, Monroe SS (2006) Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J Clin Microbiol 44(4):1405–1412
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet News 4, p 14
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
International Centre for Diarrhoeal Disease Research, Bangladesh (2006) Estimated deaths due to rotavirus in Bangladesh. Health Sci Bull 4(1):6–10
Bon F, Fascia P, Dauvergne M, Tenenbaum D, Planson H, Petion AM, Pothier P, Kohli E (1999) Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J Clin Microbiol 37(9):3055–3058
Buesa J, Collado B, López-Andújar P, Abu-Mallouh R, Rodríguez Díaz J, García Díaz A, Prat J, Guix S, Llovet T, Prats G, Bosch A (2002) Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J Clin Microbiol 40(8):2854–2859
Farkas T, Jiang X, Guerrero ML, Zhong W, Wilton N, Berke T, Matson DO, Pickering LK, Ruiz-Palacios G (2000) Prevalence and genetic diversity of human caliciviruses (HuCVs) in Mexican children. J Med Virol 62(2):217–223
Foley B, O’Mahony J, Morgan SM, Hill C, Morgan JG (2000) Detection of sporadic cases of Norwalk-like virus (NLV) and astrovirus infection in a single Irish hospital from 1996 to 1998. J Clin Virol 17(2):109–117
Hansman GS, Doan LT, Kguyen TA, Okitsu S, Katayama K, Ogawa S, Natori K, Takeda N, Kato Y, Nishio O, Noda M, Ushijima H (2004) Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch Virol 149(9):1673–1688
Kirkwood CD, Bishop RF (2001) Molecular detection of human calicivirus in young children hospitalized with acute gastroenteritis in Melbourne, Australia, during 1999. J Clin Microbiol 39(7):2722–2724
Chen SY, Chang YC, Lee YS, Chao HC, Tsao KC, Lin TY, Ko TY, Tsai CN, Chiu CH (2007) Molecular epidemiology and clinical manifestations of viral gastroenteritis in hospitalized pediatric patients in Northern Taiwan. J Clin Microbiol 45(6):2054–2057
Nayak MK, Chatterjee D, Nataraju SM, Pativada M, Mitra U, Chatterjee MK, Saha TK, Sarkar U, Krishnan T (2009) A new variant of Norovirus GII.4/2007 and inter-genotype recombinant strains of NVGII causing acute watery diarrhoea among children in Kolkata, India. J Clin Virol 45(3):223–229
Monica B, Ramani S, Banerjee I, Primrose B, Iturriza-Gomara M, Gallimore CI, Brown DW, Fathima M, Moses PD, Gray JJ, Kang G (2007) Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South India. J Med Virol 79(5):544–551
Bok K, Abente EJ, Realpe-Quintero M, Mitra T, Sosnovtsev SV, Kapikian AZ, Green KY (2009) Evolutionary dynamics of GII.4 noroviruses over a thirty-four year period. J Virol 83(22):11890–11901
Buesa J, Montava R, Abu-Mallouh R, Fos M, Ribes JM, Bartolomé R, Vanaclocha H, Torner N, Domínguez A (2008) Sequential evolution of genotype GII.4 norovirus variants causing gastroenteritis outbreaks from 2001 to 2006 in Eastern Spain. J Med Virol 80(7):1288–1295
Green KY (1997) The role of human caliciviruses in epidemic gastroenteritis. Arch Virol Suppl 13:153–165
Nakata S, Kogawa K, Numata K, Ukae S, Adachi N, Matson DO, Estes MK, Chiba S (1996) The epidemiology of human calicivirus/Sapporo/82/Japan. Arch Virol Suppl 12:263–270
Chikhi-Brachet R, Bon F, Toubiana L, Pothier P, Nicolas JC, Flahault A, Kohli E (2002) Virus diversity in a winter epidemic of acute diarrhea in France. J Clin Microbiol 40(11):4266–4272
de Wit MA, Koopmans MP, Kortbeek LM, van Leeuwen NJ, Bartelds AI, van Duynhoven YT (2001) Gastroenteritis in sentinel general practices, The Netherlands. Emerg Infect Dis 7(1):82–91
Dey SK, Nguyen TA, Phan TG, Nishio O, Salim AF, Rahman M, Yagyu F, Okitsu S, Ushijima H (2007) Molecular and epidemiological trend of norovirus associated gastroenteritis in Dhaka City, Bangladesh. J Clin Virol 40(3):218–223
Patel MM, Hall AJ, Vinjé J, Parashar UD (2009) Noroviruses: a comprehensive review. J Clin Virol 44(1):1–8
Estes MK, Prasad BV, Atmar RL (2006) Noroviruses everywhere: has something changed? Curr Opin Infect Dis 19(5):467–474
Siebenga JJ, Vennema H, Zheng DP, Vinjé J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O’Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M (2009) Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis 200(5):802–812
Victoria M, Carvalho-Costa FA, Heinemann MB, Leite JP, Miagostovich M (2007) Prevalence and molecular epidemiology of noroviruses in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil, 2004. Pediatr Infect Dis J 26(7):602–606
Okada M, Ogawa T, Kaiho I, Shinozaki K (2005) Genetic analysis of noroviruses in Chiba Prefecture, Japan, between 1999 and 2004. J Clin Microbiol 43(9):4391–4401
Gomes KA, Stupka JA, Gómez J, Parra GI (2007) Molecular characterization of calicivirus strains detected in outbreaks of gastroenteritis in Argentina. J Med Virol 79(11):1703–1709
Acknowledgments
This work was supported by the ICDDR,B and its donors, which provide unrestricted support to the Centre for its operations and research. Current donors providing unrestricted support include: Government of the People’s Republic of Bangladesh, Canadian International Development Agency (CIDA), Swedish International Development Cooperation Agency (Sida), Swiss Agency for Development and Cooperation (SDC), and Department for International Development, UK (DFID). We gratefully acknowledge these donors for their support and commitment to the Centre’s research efforts.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rahman, M., Hassan, Z., Nahar, Z. et al. Molecular detection of noroviruses in hospitalized patients in Bangladesh. Eur J Clin Microbiol Infect Dis 29, 937–945 (2010). https://doi.org/10.1007/s10096-010-0948-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-010-0948-5