Abstract
Background
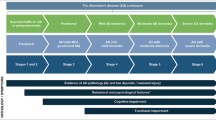
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disorder. An increasing number of researchers have found extra motor features in ALS, which are also called ALS-plus syndromes. Besides, a great majority of ALS patients also have cognitive impairment. However, clinical surveys of the frequency and genetic background of ALS-plus syndromes are rare, especially in China.
Methods
We investigated a large cohort of 1015 patients with ALS, classifying them into six groups according to different extramotor symptoms and documenting their clinical manifestations. Meanwhile, based on their cognitive function, we divided these patients into two groups and compared demographic characteristics. Genetic screening for rare damage variants (RDVs) was also performed on 847 patients.
Results
As a result, 16.75% of patients were identified with ALS-plus syndrome, and 49.5% of patients suffered cognitive impairment. ALS-plus group had lower ALSFRS-R scores, longer diagnostic delay time, and longer survival times, compared to ALS pure group. RDVs occurred less frequently in ALS-plus patients than in ALS-pure patients (P = 0.042) but showed no difference between ALS-cognitive impairment patients and ALS-cognitive normal patients. Besides, ALS-cognitive impairment group tends to harbour more ALS-plus symptoms than ALS-cognitive normal group (P = 0.001).
Conclusion
In summary, ALS-plus patients in China are not rare and show multiple differences from ALS-pure patients in clinical and genetic features. Besides, ALS-cognitive impairment group tends to harbour more ALS-plus syndrome than ALS-cognitive normal group. Our observations correspond with the theory that ALS involves several diseases with different mechanisms and provide clinical validation.



Similar content being viewed by others
References
van Es MA, Hardiman O, Chio A et al (2017) Amyotrophic lateral sclerosis. Lancet. 390:2084–2098
Chia R, Chiò A, Traynor BJ (2018) Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol 17:94–102
Ghasemi M, Brown RH Jr (2018) Genetics of amyotrophic lateral sclerosis. Cold Spring Harb Perspect Med 8:a024125
McCombe PA (2017) Wray, N R & Henderson, R D Extra-motor abnormalities in amyotrophic lateral sclerosis: another layer of heterogeneity. Expert Rev Neurother 17:561–577
Turner MR, Swash M (2015) The expanding syndrome of amyotrophic lateral sclerosis: a clinical and molecular odyssey. J Neurol Neurosurg Psychiatry 86:667–673
Brooks BR, Miller RG, Swash M et al (2000) El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 1:293–299
Blokhuis AM, Groen EJ, Koppers M et al (2013) Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol 125:777–794
Saberi S, Stauffer JE, Schulte DJ et al (2015) Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol Clin 33:855–876
Deng H, Gao K, Jankovic J (2014) The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol 10:337–348
Rademakers R, Stewart H, Dejesus-Hernandez M et al (2010) Fus gene mutations in familial and sporadic amyotrophic lateral sclerosis. Muscle Nerve 42:170–6
Cui B, Cui L, Gao J et al (2015) Cognitive Impairment in Chinese patients with sporadic amyotrophic lateral sclerosis. PloS One 10:e0137921
Niblock M, Smith BN, Lee Y-B et al (2016) Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD. Acta Neuropathol Commun 4:18
Gromicho M, Pinto S, Gisca E et al (2018) Frequency of C9orf72 hexanucleotide repeat expansion and SOD1 mutations in Portuguese patients with amyotrophic lateral sclerosis. Neurobiol Aging 70:325.e7–325.e15. https://doi.org/10.1016/j.neurobiolaging.2018.05.009 Epub 2018 May 14. 2018; 70, 325.e7-325.e15
Kanekura K, Yagi T, Cammack AJ et al (2016) Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation. Hum Mol Genet 25:1803–1813
Floris G, Borghero G, Cannas A et al (2014) Constructional apraxia in frontotemporal dementia associated with the C9orf72 mutation: broadening the clinical and neuropsychological phenotype. Amyotroph Lateral Scler Frontotemporal Degener 16:1–8
Wilke C, Pomper JK, Biskup S et al (2016) Atypical parkinsonism in C9orf72 expansions: a case report and systematic review of 45 cases from the literature. J Neurol 263:558–74
Floris G, Borghero G, Cannas A et al (2012) Frontotemporal dementia with psychosis, parkinsonism, visuo-spatial dysfunction, upper motor neuron involvement associated to expansion of C9ORF72: a peculiar phenotype? J Neurol 259:1749–1751
Geser F, Brandmeir NJ, Kwong LK et al (2008) Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Arch Neurol 65:636–641
Brettschneider J, Del Tredici K, Toledo JB et al (2013) Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol 74:20–38
Shefner JM, Al-Chalabi A, Baker MR et al (2020) A proposal for new diagnostic criteria for ALS. Clin Neurophysiol 131:1975–1978
McCluskey L, Vandriel S, Elman L et al (2014) ALS-Plus syndrome: non-pyramidal features in a large ALS cohort. J Neurol Sci 345:118–124
Gilbert RM, Fahn S, Mitsumoto H et al (2010) Parkinsonism and motor neuron diseases: twenty-seven patients with diverse overlap syndromes. Mov Disord 25:1868–1875
Cedarbaum JM, Stambler N, Malta E et al (1999) The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J Neurol Sci 169:13–21
Kimura F, Fujimura C, Ishida S et al (2006) Progression rate of ALSFRS-R at time of diagnosis predicts survival time in ALS. Neurology. 66:265–267
Liu Z, Yuan Y, Wang M et al (2021) Mutation spectrum of amyotrophic lateral sclerosis in Central South China. Neurobiol Aging 107:181–188
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164
Liao P, Yuan Y, Liu Z et al (2022) Association of variants in the KIF1A gene with amyotrophic lateral sclerosis. Transl Neurodegener 11:46
Li J, Zhao T, Zhang Y et al (2018) Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res 46:7793–7804
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Faul F, Erdfelder E, Lang AG et al (2007) G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 39:175–191
Poewe W (2008) Non-motor symptoms in Parkinson’s disease. Eur J Neurol 15:14–20
Takeda T, Kitagawa K, Arai K (2020) Phenotypic variability and its pathological basis in amyotrophic lateral sclerosis. Neuropathology. 40:40–56
Hammad M, Silva A, Glass J et al (2007) Clinical, electrophysiologic, and pathologic evidence for sensory abnormalities in ALS. Neurology. 69:2236–2242
Sakamoto H, Akamatsu M, Hirano M et al (2014) Multiple system involvement in a Japanese patient with a V31A mutation in the SOD1 gene. Amyotroph Lateral Scler Frontotemporal Degener 15:312–314
Kawata A, Kato S, Hayashi H et al (1997) Prominent sensory and autonomic disturbances in familial amyotrophic lateral sclerosis with a Gly93Ser mutation in the SOD1 gene. J Neurol Sci 153:82–85
Giannini F, Battistini S, Mancuso M et al (2010) D90A-SOD1 mutation in ALS: the first report of heterozygous Italian patients and unusual findings. Amyotroph Lateral Scler 11:216–219
Marjanovic IV, Selak-Djokic B, Peric S et al (2017) Comparison of the clinical and cognitive features of genetically positive ALS patients from the largest tertiary center in Serbia. J Neurol 264:1091–1098
Pradat PF, Bruneteau G, Munerati E et al (2009) Extrapyramidal stiffness in patients with amyotrophic lateral sclerosis. Mov Disord 24:2143–2148
Calvo A, Chiò A, Pagani M et al (2019) Parkinsonian traits in amyotrophic lateral sclerosis (ALS): a prospective population-based study. J Neurol 266:1633–1642
Nishihira Y, Tan C-F, Onodera O et al (2008) Sporadic amyotrophic lateral sclerosis: two pathological patterns shown by analysis of distribution of TDP-43-immunoreactive neuronal and glial cytoplasmic inclusions. Acta Neuropathol 116:169
Merner ND, Girard SL, Catoire H et al (2012) Exome sequencing identifies FUS mutations as a cause of essential tremor. Am J Hum Genet 91:313–319
Corrado L, Del Bo R, Castellotti B et al (2010) Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J Med Genet 47:190–194
Fujita K, Matsubara T, Miyamoto R et al (2019) Co-morbidity of progressive supranuclear palsy and amyotrophic lateral sclerosis: a clinical-pathological case report. BMC Neurol 19:168
Nagao S, Yokota O, Nanba R et al (2012) Progressive supranuclear palsy presenting as primary lateral sclerosis but lacking parkinsonism, gaze palsy, aphasia, or dementia. J Neurol Sci 323:147–153
Goldman J, Waters C, Mitsumoto H et al (2014) Multiple system atrophy and amyotrophic lateral sclerosis in a family with hexanucleotide repeat expansions in C9orf72 (P4.105). Neurology. 82:P4.105
Wenning GK, Tison F, Ben Shlomo Y et al (1997) Multiple system atrophy: a review of 203 pathologically proven cases. Mov Disord 12:133–147
Chiò A, Logroscino G, Hardiman O et al (2009) Prognostic factors in ALS: a critical review. Amyotroph Lateral Scler 10:310–323
Mizuno Y (2004) Progress in the basic and clinical aspects of Parkinson’s disease. Rinsho Shinkeigaku 44:741–750
Su WM, Cheng YF, Jiang Z et al (2021) Predictors of survival in patients with amyotrophic lateral sclerosis: a large meta-analysis. EBioMedicine. 74:103732
Punjani R, Larson TC, Wagner L et al (2018) Survival and epidemiology of amyotrophic lateral sclerosis (ALS) cases in the Chicago and Detroit metropolitan cohort: incident cases 2009-2011 and survival through. Amyotroph Lateral Scler Frontotemporal Degener 2022:1–9
Ringholz GM, Appel SH, Bradshaw M et al (2005) Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 65:586–590
Bergner CG, Neuhofer CM, Funke C et al (2020) Case report: association of a variant of unknown significance in the FIG4 gene with frontotemporal dementia and slowly progressing motoneuron disease: a case report depicting common challenges in clinical and genetic diagnostics of rare neuropsychiatric and neurologic disorders. Front Neurosci 14:559670
Chung EJ, Hwang JH, Lee MJ et al (2014) Expansion of the clinicopathological and mutational spectrum of Perry syndrome. Parkinsonism Relat Disord 20:388–393
Milanowski L, Sitek EJ, Dulski J et al (2020) Cognitive and behavioral profile of Perry syndrome in two families. Parkinsonism Relat Disord 77:114–120
Münch C, Sedlmeier R, Meyer T et al (2004) Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology. 63:724–726
Liu X, Yang L, Tang L et al (2017) DCTN1 gene analysis in Chinese patients with sporadic amyotrophic lateral sclerosis. PloS One 12:e0182572
Broustal O, Camuzat A, Guillot-Noël L et al (2010) FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J Alzheimers Dis 22:765–769
Nakamura M, Bieniek KF, Lin WL et al (2015) A truncating SOD1 mutation, p.Gly141X, is associated with clinical and pathologic heterogeneity, including frontotemporal lobar degeneration. Acta Neuropathol 130:145–157
Olney RK, Murphy J, Forshew D et al (2005) The effects of executive and behavioral dysfunction on the course of ALS. Neurology. 65:1774–1777
Wolf J, Safer A, Wohrle JC et al (2014) Variability and prognostic relevance of different phenotypes in amyotrophic lateral sclerosis - data from a population-based registry. J Neurol Sci 345:164–167
Yang T, Hou Y, Li C et al (2021) Risk factors for cognitive impairment in amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 92:688–693
Nakayama Y, Shimizu T, Matsuda C et al (2018) Non-motor manifestations in ALS patients with tracheostomy and invasive ventilation. Muscle Nerve 57:735–741
Dewan R, Chia R, Ding J et al (2021) Pathogenic Huntingtin repeat expansions in patients with frontotemporal dementia and amyotrophic lateral sclerosis. Neuron. 109:448–460.e4
Acknowledgements
We are grateful to the participating patients for their involvement. We would like to thank the patients for permitting us to publish this information.
Data sharing and data accessibility
The original contributions presented in the study are included in the article/Supplementary material; further inquiries can be directed to the corresponding authors.
Funding
This work was supported by the Science and Technology Innovation 2030 (STI2030-Major Projects:2021ZD0201803 to J. W.); the National Key R&D Program of China (#2021YFA0805202 and #2018YFC1312003 to J. W; 2018YFC2000300 to X. Z); National Natural Science Foundation of China (#82171431, 81671120 and 81300981 to J. W.); the Natural Science Fund for Distinguished Young Scholars of Hunan Province, China (#2020JJ2057 to J. W.) and the Project Program of National Clinical Research Center for Geriatric Disorders at Xiangya Hospital (#2020LNJJ13 to J. W.).
Author information
Authors and Affiliations
Contributions
C. C., Q. Z., P. L., L. S., J. G., B. T., J. W. and X. Z. contributed to the conception and design of the study; X. T., Z. L., W. L., Y. H., H. J., Y. Y., X. H. and B. J. contributed to the acquisition and analysis of the data; C. C., Q. Z., P. L., Y. Y., Z. L., X. Z. and J. W. contributed to the drafting of the manuscript and in preparing the figures.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from all the participants and the study protocol was approved by the Ethics Committee and the Expert Committee of Xiangya Hospital, Central South University; the number of the approval is 202106134.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary file 1
Supplemental Figure 1 The frequency of different gene mutations in the ALS-pure or ALS-plus group.
Supplementary file 2
Supplemental Figure 2 The frequency of different gene mutations in the ALS-cognitive normal or ALS-cognitive impairment group.
Supplementary file 3
Supplemental Table 1 Logistic analysis between the ALS-pure and ALS-plus groups.
Supplementary file 4
Supplemental Table 2 RDVs and demographic characteristics in the ALS-pure and ALS-plus groups.
Supplementary file 5
Supplemental Table 3 Rare disease variants (RDVs) of PD-related gene in the ALS-pure and ALS-plus groups.
Supplementary file 6
Supplemental Table 4 Ratio of comorbidity among ALS subgroups.
Supplementary file 7
Supplemental Table 5 Cox analysis about factors associated with survival in ALS patients
Supplementary file 8
Supplemental Table 6 RDVs and demographic characteristics in the ALS-cognitive impairment and ALS-cognitive normal groups.
Supplementary file 9
Supplemental Table 7 Logistic analysis between the ALS-cognitive impairment and ALS-cognitive normal groups.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chang, C., Zhao, Q., Liu, P. et al. ALS-plus related clinical and genetic study from China. Neurol Sci 44, 3557–3566 (2023). https://doi.org/10.1007/s10072-023-06843-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-023-06843-4