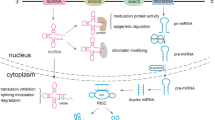
Abstract
Multiple sclerosis (MS) as chronic autoimmune inflammatory neurological disease of the central nervous system (CNS) occurs due to several environmental and genetic factors, whose pathogenesis is associated with genes with regulatory role in the immune system. Long non-coding RNAs (LncRNAs) are able to reportedly regulate responses of immune systems and expression of genes, and show the tissue specificity and complexity of biofunctions. Various studies have suggested that the aberrant LncRNA expression is an underlying factor involved in the incidence of MS and that the analysis of the expression profile of these molecules can be a specific biomarker of MS for preventing the course of the disease or responding to treatment. The purpose of this research was to review the recent studies for exploring the functions of LncRNAs in the processes leading to MS disease.


Similar content being viewed by others
References
Lublin FD (2014) New multiple sclerosis phenotypic classification. Eur Neurol 72(Suppl. 1):1–5
Soudyab M, Iranpour M, Ghafouri-Fard S (2016) The role of long non-coding RNAs in breast cancer. Arch Iran Med (AIM) 19(7):508–517
Wu G-C, Pan H-F, Leng R-X, Wang D-G, Li X-P, Li X-M, Ye DQ (2015) Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev 14(9):798–805
Cao J (2014) The functional role of long non-coding RNAs and epigenetics. Biol Proced Online 16(1):42
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10(3):155–159
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47(3):199–208
Spurlock CF 3rd, Crooke PS 3rd, Aune TM (2016) Biogenesis and transcriptional regulation of long noncoding RNAs in the human immune system. J Immunol (12):19717–14509
Yu AD, Wang Z, Morris KV (2015) Long noncoding RNAs: a potent source of regulation in immunity and disease. Immunol Cell Biol 93(3):277–283
Kaikkonen MU, Lam MT, Glass CK (2011) Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 90(3):430–440
Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature. 482(7385):339–346
Mattick JS, Rinn JL (2015) Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol 22(1):5–7
Lam MT, Li W, Rosenfeld MG, Glass CK (2014) Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39(4):170–182
Ledford H (2013) Circular RNAs throw genetics for a loop. Nature. 494(7438):415
Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA et al (2010) Expression of distinct RNAs from 3′ untranslated regions. Nucleic Acids Res 39(6):2393–2403
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 316(5830):1484–1488
Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell. 152(6):1298–1307
Mattick JS (2009) The genetic signatures of noncoding RNAs. PLoS Genet 5(4):e1000459
Gugnoni M, Ciarrocchi A (2019) Long noncoding RNA and epithelial mesenchymal transition in cancer. Int J Mol Sci 20(8):1924
Tano K, Akimitsu N (2012) Long non-coding RNAs in cancer progression. Front Genet 3:219
Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Lis RT, Zhao SG, Wu Q, Feng FY, Loda M, He HH, Liu XS, Brown M (2017) Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U S A 114(26):E5207–E5e15
Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM (2014) The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 111(3):1002–1007
Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K (2013) Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 14(11):1190–1198
Tong Q, Gong AY, Zhang XT, Lin C, Ma S, Chen J, Hu G, Chen XM (2016) LincRNA-Cox2 modulates TNF-alpha-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J 30(3):1187–1197
Yang X, Yang J, Wang J, Wen Q, Wang H, He J et al (2016) Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Sci Rep 6:38963
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X (2014) The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 344(6181):310–313
Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT (2008) Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5). J Cell Sci 121(7):939–946
Mourtada-Maarabouni M, Hasan AM, Farzaneh F, Williams GT (2010) Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Mol Pharmacol 78(1):19–28
Williams GT, Mourtada-Maarabouni M, Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Portland Press Limited; 2011
Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K (2013) The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 152(4):743–754
Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM (2012) Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol (Baltimore, Md : 1950) 189(5):2084–2088
Vigneau S, Rohrlich PS, Brahic M, Bureau JF (2003) Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J Virol 77(10):5632–5638
Collier SP, Henderson MA, Tossberg JT, Aune TM (2014) Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol 193(8):3959–3965
Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S et al (2015) The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 16(3):318–325
Petri A, Dybkaer K, Bogsted M, Thrue CA, Hagedorn PH, Schmitz A et al (2015) Long noncoding RNA expression during human B-cell development. PLoS One 10(9):e0138236
Sehgal L, Mathur R, Braun FK, Wise JF, Berkova Z, Neelapu S, Kwak LW, Samaniego F (2014) FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia. 28(12):2376–2387
Kunz M, Ibrahim SM (2009) Cytokines and cytokine profiles in human autoimmune diseases and animal models of autoimmunity. Mediat Inflamm 2009:1–20
Thompson AJ, Baranzini SE, Geurts J, Hemmer B, Ciccarelli O (2018) Multiple sclerosis. Lancet (London, England) 391(10130):1622–1636
Mahad DH, Trapp BD, Lassmann H (2015) Pathological mechanisms in progressive multiple sclerosis. Lancet Neurol 14(2):183–193
Confavreux C, Vukusic S (2014) The clinical course of multiple sclerosis. Handb Clin Neurol 122:343–369
Santoro M, Nociti V, Lucchini M, De Fino C, Losavio FA, Mirabella M (2016) Expression profile of long non-coding RNAs in serum of patients with multiple sclerosis. J Mol Neurosci 59(1):18–23
Zhang F, Liu G, Bu Y, Ma X, Hao J (2016) Expression profile of long noncoding RNAs and mRNAs in peripheral blood mononuclear cells from myasthenia gravis patients. J Neuroimmunol 299:124–129
Eftekharian MM, Ghafouri-Fard S, Soudyab M, Omrani MD, Rahimi M, Sayad A, Komaki A, Mazdeh M, Taheri M (2017) Expression analysis of long non-coding RNAs in the blood of multiple sclerosis patients. J Mol Neurosci 63(3–4):333–341
Li Z, Chao T-C, Chang K-Y, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM (2014) The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci 111(3):1002–1007
Kalliolias GD, Ivashkiv LB (2016) TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol 12(1):49–62
Colombo T, Farina L, Macino G, Paci P (2015) PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int 2015:1–10
Aune TM, Spurlock CF III (2016) Long non-coding RNAs in innate and adaptive immunity. Virus Res 212:146–160
Cardamone G, Paraboschi EM, Rimoldi V, Duga S, Soldà G, Asselta R (2017) The characterization of GSDMB splicing and backsplicing profiles identifies novel isoforms and a circular RNA that are dysregulated in multiple sclerosis. Int J Mol Sci 18(3):576
Iparraguirre L, Muñoz-Culla M, Prada-Luengo I, Castillo-Triviño T, Olascoaga J, Otaegui D (2017) Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum Mol Genet 26(18):3564–3572
Turner M, Galloway A, Vigorito E (2014) Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol 15(6):484–491
Kipanyula MJ, Kimaro WH, Etet PF (2016) The emerging roles of the calcineurin-nuclear factor of activated T-lymphocytes pathway in nervous system functions and diseases. J Aging Res 2016:1–20
Fenoglio C, Oldoni E, Serpente M, De Riz MA, Arcaro M, D'Anca M et al (2018) LncRNAs expression profile in peripheral blood mononuclear cells from multiple sclerosis patients. J Neuroimmunol 324:129–135
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477(7364):295
Rossi S, Motta C, Studer V, Macchiarulo G, Volpe E, Barbieri F, Ruocco G, Buttari F, Finardi A, Mancino R, Weiss S, Battistini L, Martino G, Furlan R, Drulovic J, Centonze D (2014) Interleukin-1β causes excitotoxic neurodegeneration and multiple sclerosis disease progression by activating the apoptotic protein p53. Mol Neurodegener 9(1):56
Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF (2010) Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci 11(1):14
Barry G, Briggs J, Vanichkina D, Poth E, Beveridge N, Ratnu V et al (2014) The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry 19(4):486–494
Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, Hussain MS, Nejad P, Patel B, Hei H, Khoury S, Quintana F, Kivisakk P, Chitnis T, Weiner HL (2013) Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol 73(6):729–740
Tehrani SS, Karimian A, Parsian H, Majidinia M, Yousefi B (2018) Multiple functions of long non-coding RNAs in oxidative stress, DNA damage response and cancer progression. J Cell Biochem 119(1):223–236
Dastmalchi R, Omrani MD, Mazdeh M, Arsang-Jang S, Movafagh A, Sayad A et al (2018) Expression of long non-coding RNAs (UCA1 and CCAT2) in the blood of multiple sclerosis patients: a case - control study. Iran Red Crescent Med J 20(8):e66334
Xue Y, Teng Y-Q, Zhou J-D, Rui Y-J (2016) Long noncoding RNA CCAT2 as an independent prognostic marker in various carcinomas: evidence based on four studies. Int J Clin Exp Med 9(2):2567–2572
Ong MS, Cai W, Yuan Y, Leong HC, Tan TZ, Mohammad A, You ML, Arfuso F, Goh BC, Warrier S, Sethi G, Tolwinski NS, Lobie PE, Yap CT, Hooi SC, Huang RY, Kumar AP (2017) ‘Lnc’-ing Wnt in female reproductive cancers: therapeutic potential of long non-coding RNAs in Wnt signalling. Br J Pharmacol 174(24):4684–4700
Dastmalchi R, Ghafouri-Fard S, Omrani MD, Mazdeh M, Sayad A, Taheri M (2018) Dysregulation of long non-coding RNA profile in peripheral blood of multiple sclerosis patients. Mult Scler Relat Disord 25:219–226
Abedin K, Papadopoulos D, Dexter D, Reynolds R (2002) Oxidative DNA damage in the spinal cord in multiple sclerosis. Neuropathol Appl Neurobiol 28(2):168
Kim C, Kang D, Lee EK, Lee J-S (2017) Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxidative Med Cell Longev 2017:1–21
Ladiwala U, Li H, Antel JP, Nalbantoglu J (1999) p53 induction by tumor necrosis factor-α and involvement of p53 in cell death of human oligodendrocytes. J Neurochem 73(2):605–611
Wosik K, Antel J, Kuhlmann T, Brück W, Massie B, Nalbantoglu J (2003) Oligodendrocyte injury in multiple sclerosis: a role for p53. J Neurochem 85(3):635–644
Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel Growth. Novelty and significance. Circ Res 114(9):1389–1397
Cardamone G, Paraboschi EM, Soldà G, Cantoni C, Supino D, Piccio L, Duga S, Asselta R (2018) Not only cancer: the long non-coding RNA MALAT1 affects the repertoire of alternatively spliced transcripts and circular RNAs in multiple sclerosis. Hum Mol Genet 28(9):1414–1428
Acknowledgments
The authors would like to thank all colleagues in the Department of Medical Genetics, Faculty of Medicine, Mashhad University of Medical Sciences.
Contributorship statement
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Taghizadeh, E., Taheri, F., Samadian, M.M. et al. Role of long non-coding RNAs (LncRNAs) in multiple sclerosis: a brief review. Neurol Sci 41, 2443–2451 (2020). https://doi.org/10.1007/s10072-020-04425-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-020-04425-2